|
 |
| Organoid > Volume 4; 2024 > Article |
|
Abstract
Background
Methods
NOTES
Funding
The work was supported by a National Research Foundation (NRF) of Korea grant funded by the Korean government (MSIT) (No. NRF-2018R1D1A1A02047589), Basic Science Research Program through the National Research Foundation of Korea funded by the Ministry of Science, ICT and Future Planning, Republic of Korea (NRF-2018R1D1A102050030); the Korea Health Technology R&D Project through the Korea Health Industry Development Institute, funded by the Ministry of Health & Welfare, Republic of Korea (HI16C0002, HI17C2094, HI18C2458); and the Technology Innovation Program (or Industrial Strategic Technology Development Program 3D-TissueChip Based Drug Discovery Platform Program) (20009773, Commercialization of 3D Multifunction Tissue Mimetics Based Drug Evaluation Platform) funded by the Ministry of Trade, Industry & Energy (MOTIE, Korea).
AuthorsŌĆÖ contributions
Conceptualization: WHC, JY, HYJ; Data curation: LSJ, JHJ; Funding acquisition: JY, HYJ; Methodology: JHJ, HL; Project administration: JY, KJL; Software: JHJ, LJ; Supervision: HYJ; Validation: LJ; Visualization: SJL; Writing-original draft: KJL; Writing-review & editing: all authors.
Fig.┬Ā1.
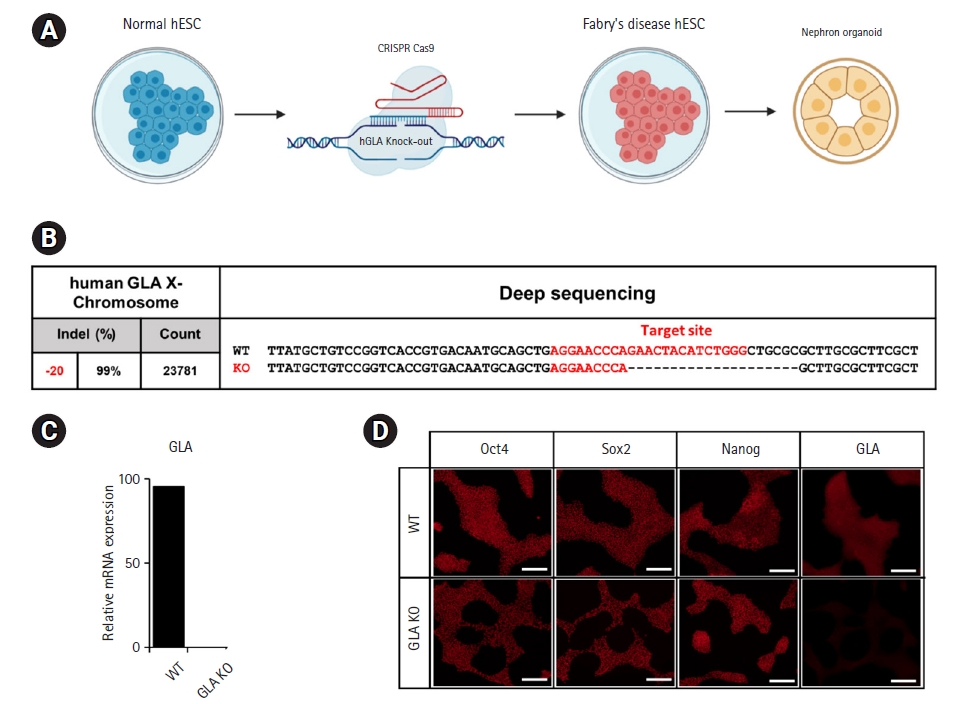
Fig.┬Ā2.

References
- TOOLS
-
METRICS

-
- 0 Crossref
- 0 Scopus
- 977 View
- 12 Download
- ORCID iDs
-
Woo Hee Choi

https://orcid.org/0000-0002-5454-2943 - Related articles



 PDF Links
PDF Links PubReader
PubReader ePub Link
ePub Link Full text via DOI
Full text via DOI Download Citation
Download Citation Print
Print