Optimizing a three-dimensional spheroid clearing method for the imaging-based evaluation of cardiotoxicity
Article information
Abstract
Background
Toxicity evaluation based on two-dimensional cell culture shows differences from clinical results and has the disadvantage of not accurately reflecting cell-to-cell cross-signaling. Since almost all cells in the human body are arranged in a three-dimensional structure and constitute a tissue, the in vitro reproduction of a three-dimensional tissue composed of human cells can be used as an effective model for drug development and toxicity evaluation. The clearing technique improves image resolution and can implement three-dimensional bio-images throughout the organization, enabling more efficient toxicity evaluation of disease model analysis using spheroids.
Methods
We generate an 100-200um spheroids and optimized clearing condition. Next application and imaging of optimized clearing condition in the cardiac spheroid model.
Results
Here we first reported that the optical spheroid clearing protocol for the image-based toxicity prediction model. In our results, spheroid clearing significantly increases fluorescence intensity and enables image-based toxicity prediction.
Conclusion
We propose that these clearing methods of spheroid can be utilized for the image-based cardiotoxicity evaluation. Furthermore, we also present the possibility that our protocol can also be utilized for patient-tailored toxicity prediction evaluation.
Introduction
Two-dimensional (2D) cell culture systems, which are in widespread general use, have the disadvantage of not fully incorporating information in vivo [1]. In contrast, 3-dimensional (3D) cell culture systems can be used to implement a more realistic biological environment that includes paracrine signaling, cell secretion diffusion, and cell-cell interactions involving extracellular substrates [2,3]. Since almost all cells of the human body are arranged in a 3D structure and constitute tissues, the reproduction of 3D tissues in vitro is an essential research field that can contribute to a deeper understanding of the physiology of human tissues and the development of new drugs. A recent study reported that stem cell-derived 3D cells exhibited more sensitive toxic reactivity than 2D-cultured cells [4]. As such, 3D cell culture systems are more similar to in vivo conditions than 2D cell culture systems, and they can be applied as useful models for toxicity prediction in new drug development.
For drug development, cell-based screening and toxicity assays have been used for several decades [5]. Cardiotoxicity is a serious form of toxicity and a very important moderator that should be considered in the drug discovery process [6]. Safety assays in active use for evaluating pharmacological cardiotoxicity include arrhythmia prediction through the in vitro human ether-a-go-go related gene (hERG) assay and methods to evaluate effects on cardiac action potential ion channels, action potentials in heart tissue in laboratory animals, and toxicity using human ventricular myocytes/stem cell-derived human cardiomyocytes (CMs), as well as in silico cardiotoxicity evaluation methods [7-9]. The hERG test is mainly used as a method to assess cardiovascular effects during essential tests of pharmacological safety, and although the sensitivity of the hERG channel to drugs is excellent, the effects of a new drug on the hERG channel may not be predictive of actual clinical/nonclinical tests [7]. Evaluations using experimental animals pose ethical problems and errors due to interspecies differences from humans. In addition, since toxicity evaluation methods using human myocardial cells are limited in supply and difficult to use, evaluations using human pluripotent stem cell (hPSC)-derived myocardial cells have been proposed as an alternative [10,11]. hPSC-derived CMs (hPSC-CMs) are similar to adult myocardial cells in terms of the expression of genes involved in ion channels in the heart and genes that are expressed specifically in myocardial cells. For this reason, studies on drug impact assessment and toxicity prediction have reported measurements of the action potential amplitude, maximum diastolic potential, and the maximum rate of rise of the action potential upstroke in ventricular-like cells to verify the electrophysiological properties of hPSC-CMs [12,13]. Recent research aiming to improve the toxicity of prediction models has utilized 3D cell culture technology of hPSC-CMs to reflect 3D structures and intercellular characteristics in vivo [1]. However, the remaining limitations of toxicity evaluation through spheroid imaging include the fact that this method does not reflect the structure of cells or marker expression in all cells.
Tissue clearing (TC) techniques, which have been actively researched recently [14-19], provide 3D morphological examination; furthermore, staining intracellular and cytoplasmic organs allows more information to be obtained, leading to reliable results and accurate predictions in toxicity evaluation. The clearing technologies reported to date include the benzyl alcohol/benzyl benzoate (BABB) method, the immunolabeling-enabled 3D imaging of solvent cleared organs (iDISCO) method, the Scale S method using organic solvents, and the active CLARITY technology (ACT) method, which involves polymer injection [14-19]. Previously, to observe the 3D structure of spheroids, the tissue was cut and made into 2D slides, and then images with micrometer-level thickness information were obtained. The acquisition of thicker intra-organizational information requires a series of processes to produce continuous intercepts and reconstructions after each image. Clearing techniques provide access to the integrated structure and molecular information by verifying the internal structure and protein distribution without tissue damage [20]. Although research on clearing is actively being conducted, most of the intensively-studied clearing techniques are focused on neuroscience and brain development [19,21]. Furthermore, although research on cancer spheroid clearing for drug screening has been reported, no stem cell-based clearing study has yet been conducted for toxicity evaluation [22,23]. In this study, we established a protocol that enables image-based cardiotoxicity prediction by incorporating clearing techniques into human stem cell-derived spheroids and suggest a new toxicity evaluation method.
Materials and Methods
Ethics statement: Ethical approval for this study was obtained from the Public Institutional Review Board Designated by Ministry of Health and Welfare, Korea (P01-102104-41-002).
Cell culture and differentiation of hPSCs
Cells from an hPSC line (H1; WiCell Research Institute, Madison, WI, USA) were seeded on matrigel (Corning Inc., Corning, NY, USA)-coated (1:200) polystyrene plates and maintained in Essential Eight medium (E8; Stemcell Technologies, Vancouver, BC, Canada). Cells were dissociated in phosphate-buffered saline (PBS) with 0.5 mM EDTA (Gibco; Thermo Fisher, Waltham, MA, USA) for 3 to 5 minutes at 37°C. For the first 24 hours after passaging, the ROCK inhibitor Y-27632 (Stemgent, Cambridge, MA, USA) was added at a concentration of 10 µM to the hPSCs’ maintenance medium. To prepare for cell differentiation, human embryonic stem cells (hESCs) were seeded on matrigel-coated plates and cultured in mTeSR-1 (Stemcell Technologies) medium until 80% to 90% confluence. To initiate cell differentiation, the media was changed to RPMI-1640 (Gibco) supplemented with B27 minus insulin (Invitrogen; Thermo Fisher). On day 0, the cells were treated with 5 µM CHIR99021. Between days 3 and 4, cells were treated with 5 µM IWP4 (Stemgent). Starting on day 7, the medium was replaced with RPMI-1640 supplemented with B27 (Invitrogen) every 2 days. To purify hPSC-CMs, the medium was replaced with glucose-deprived RPMI-1640 supplemented with 5 mM lactate (Sigma-Aldrich, St. Louis, MO, USA) for 7 days. After the purification of myocytes, cells were dissociated into single cells with 0.05% trypsin-EDTA (Gibco) and TrypLE Express (Gibco) at 37°C for 5 minutes. Cells were collected with RPMI-1640 supplementation with 10% fetal bovine serum (Gibco), and gently centrifuged at 1,000 rpm for 3 minutes. hESC-derived CMs (hESC-CMs) were replated on matrigel-coated plates with RPMI-1640 supplementation with B27.
Generation of spheroids
To establish the clearing conditions, we generated spheroids using HEK293 cell lines. The HEK293 cells were maintained in DMEM (Gibco) and the medium was changed every 48 hours. To generate spheroids, 1 × 106 cells (HEK293) were prepared and seeded on StemFIT 3D (Microfit, Seongnam, Korea). Before cell seeding, the StemFIT 3D plate was washed with 70% ethanol to prevent contamination and remove bubbles. After 24 hours, spheroids were generated in single wells. Single spheroids were transferred to vitronectin (Stemcell Technologies)-coated 96-well U-bottom plates (Greiner, Frickenhausen, Germany).
Generation of cardio-spheroids
To generate cardio-spheroids, hPSCs were differentiated into CMs as previously described. Between days 7 and 10, beating cells could be observed under microscopy. After the purification step, hPSC-CMs were treated with TrypLE Express and 0.05% trypsin-EDTA for 5 minutes at 37℃. Preparations of single-dissociated cells (1 × 106 cells) were seeded on a StemFIT 3D device. The hPSC-CMs were replated in RPMI-1640 + 1X B27 supplement (Gibco) medium containing 10 µM ROCK inhibitor Y-27632. Beating cardio-spheroids were generated after 24 to 48 hours. After spheroid generation, single spheroids were transferred to a vitronectin-coated 96-well plate for the toxicity evaluation.
Clearing and high-throughput imaging of spheroids
Spheroids were fixed in 4% paraformaldehyde at 4°C overnight and washed with PBS. They were covered with 50 µL of 0.5% agarose (Invitrogen) to fix them in position. The spheroids in 0.5% agarose were solidified at room temperature (RT) for 1 hour. The spheroids were incubated overnight in 200 µL of TC solution at 37°C under shaking conditions. The TC solution was composed of 25 w/v% compound C (Detergent Mixture; Binaree, Daegu, Korea) and 50% urea (Junsei, Tokyo, Japan) in distilled water (DW). The solution was stored at RT and pre-warmed at 37°C until used. The spheroids underwent rinsing and washing in DW, and were then incubated for a second time in TC solution under the same conditions. After washing with DW, the spheroids were incubated in 0.1 × PBS with 0.2 w/v% Triton X-100 and 10% dimethyl sulfoxide (permeabilization solution) at 4°C for 2 days. The primary antibody was diluted in 0.1 × PBS with 0.01% sodium azide and 0.1% Tween20 (antibody dilution solution) at a dilution of 1:100. The spheroids were incubated with a primary antibody (CnT, pro-BNP, a-actinin, tubulin; Abcam, Cambridge, UK; α-actinin; Abcam, tubulin; Abcam) for 2 days at 4°C. Next, the spheroids were washed 5 times in DW, and then the spheroids were incubated with secondary fluorescence-conjugated antibodies for 2 days. After being washed with DW 5 times, nuclear staining was performed with DAPI at RT for 1 hour. The spheroids were then washed with DW and mounted with mounting solution (C 40 w/v%, urea 40 w/v%, in 0.1 × PBS) at 37°C under shaking conditions. Samples were stored at RT until they were taken for imaging. After clearing, 96-well plates with spheroids were imaged on an Image Xpress Micro XLS high-content microscope (Molecular Devices, San Jose, CA, USA).
Statistical analysis
The statistical analysis was conducted using GraphPad Prism 5 (GraphPad Inc., La Jolla, CA, USA). Data are expressed as the mean ± standard error of the mean. Statistical significance was determined by the Student t-test for comparisons between 2 groups.
Results
Scheme for optimized spheroid clearing
To optimize spheroid clearing, we prepared spheroids using HEK-293 cell lines. We generated spheroids with an average diameter of 100 to 200 µm using StemFIT 3D. To prevent the spheroids from moving, they were transferred into a vitronectin-coated 96-well plate and fixed with agarose for use in this experiment. Although the data are not shown, the efficiency was analyzed by comparing vitronectin, matrigel, and gelatin as coating materials for spheroid fixation; vitronectin, as the most adhesive substance, was selected and used for the experiment (coating for over 1 hour). Next, 50 µL/well of 0.5% agarose was added for spheroid position fixation. In agarose solutions with concentrations over 0.6%, the clearing solution did not penetrate the spheroid, affecting the efficiency of the clearing. First, the clearing solution was optimized in terms of the number of exposure solutions and incubation times, and the optical clearing of the spheroids was checked. Next, immunofluorescence optimization was performed. Clearing efficiency was confirmed by optical clearing of bright-field images and mean fluorescence intensity. Finally, we selected the optimized clearing conditions and acquired fluorescence images of cardio-spheroids (Fig. 1).
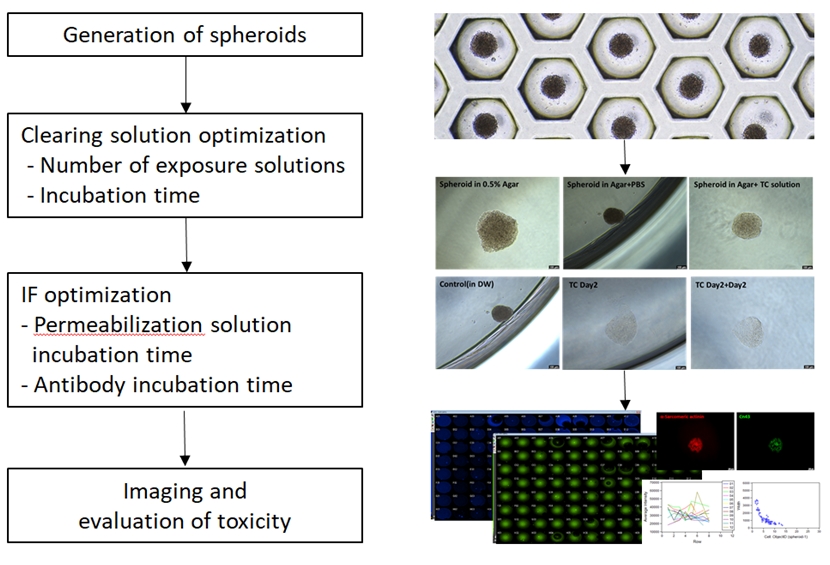
Schematic illustration of optimized spheroid clearing. To establish spheroid clearing conditions, the first step was to generate spheroids. The next step was to investigate the number of clearing solution exposures and incubation time. Subsequently, the immunofluorescence (IF) conditions were determined by optimizing the permeabilization solution and antibody incubation time. Finally, optimized clearing conditions were applied to spheroids for imaging and toxicity evaluation. PBS, phosphate-buffered saline; TC, tissue clearing; DW, distilled water.
Optimization of spheroid clearing conditions
We generated spheroids using HEK293 cell lines and incubated them in different numbers of TC solutions for different time periods to optimize the spheroid clearing method. First, we compared and verified the degree of clearing according to the number of TC solution treatments. The spheroids were fixed and incubated with different numbers of TC solutions, and we acquired bright-field images of the spheroids after clearing. Spheroid cleaning drastically improved as the number of TC solutions increased (Fig. 2A). The bright-field images revealed that the clearing of the spheroids progressed the most with 2 treatments of the TC solution and culture for 3 days (Fig. 2A). To determine the most appropriate clearing solution incubation time, the spheroids were incubated with TC solution for various time intervals and imaged with light microscopy. Long-term incubation with a clearing solution improved the clearing of spheroids (Fig. 2B). Next, to address the effects of TC solution incubation time on fluorescence, we stained spheroids with tubulin (green) and DAPI (blue). We found that the spheroids that were incubated twice with 3 days of incubation in TC solution showed the highest fluorescence intensities. Based on the results, we selected 2 treatments and 3 days of incubation in TC solution to optimize spheroid clearing.

Establishment of spheroid clearing conditions. (A) Bright-field images of spheroids (HEK293-derived spheroids) with different numbers of incubations in tissue clearing (TC) solution. Non-incubated clearing solution (0), 1 (1 time), 2 (TC2, 2 times), 2 (TC3, 2 times and 3 days of incubation in the TC solution); scale bar, 100 µm. (B) Analysis of clearing efficacy based on clearing solution incubation times using bright-field acquisitions; scale bar, 100 µm. (C) Representative fluorescence images and fluorescence quantitation graphs of treated spheroids with different numbers of TC solutions; scale bar, 100 µm. Data are represented as mean ± standard error of the mean.
*** p < 0.001 vs. Non-incubated clearing solution (0).
To determine the best conditions for permeabilization, 0.2% Triton X-100 solution was used for spheroid fixation. We proceeded with the clearing of the spheroids based on the previously obtained clearing conditions and compared the fluorescence intensity through tubulin staining. As the exposure time in the permeabilization solution increased, the mean fluorescence intensity significantly increased (Fig. 3A). The intensity of spheroids exposed to solutions for 3 days was somewhat higher than that of those exposed for 2 days, but there was no significant difference. Based on these results, 2 days was selected as the incubation time of the permeabilization solution.
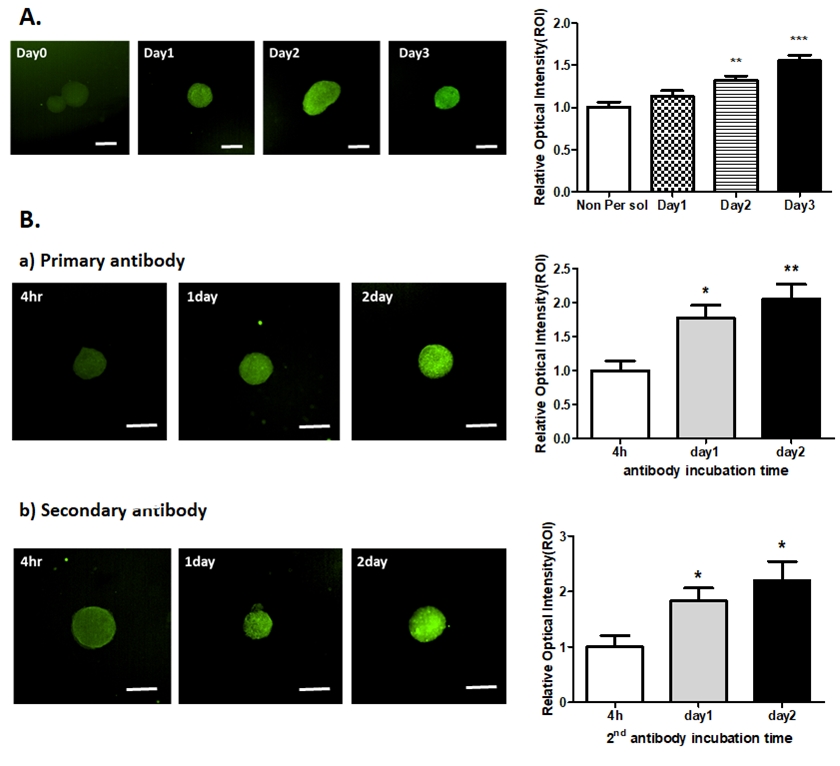
Establishment of spheroid permeabilization conditions. (A) Different permeabilization times for the acquisition of optimized fluorescence images (HEK293-derived spheroids); scale bar, 100 µm. **p<0.01, ***p<0.001 vs. day0, (B) Comparision of the antibody incubation time for the acquisition of optimized fluorescence images. Upper panel: representative images of different times of primary antibody incubation, lower panel: representative images of different times of secondary antibody incubation; scale bar, 100 µm. Data are represented as mean ± standard error of the mean. *p<0.05, **p<0.01 vs. 4h.
Next, an experiment was conducted to compare the fluorescence intensity according to the antibody incubation time. As the exposure time in the antibody incubation increased, the mean fluorescence intensity significantly increased (Fig 3B). Based on this result, we selected 2 days as the antibody incubation time to optimize spheroid clearing.
Application and imaging of optimized clearing conditions in the cardio-spheroid model
Based on the previous data, we established a protocol optimized for cardio-spheroid clearing as follows (Fig. 4A). Cardio-spheroids were generated and fixed with 4% paraformaldehyde at 4°C overnight. The spheroids were solidified in 0.5% agarose at RT for 1 hour. Then, the cardio-spheroids were incubated overnight in a clearing solution at 37℃ under conditions of shaking. After incubation and washing with DW, the spheroids were incubated a second time with clearing solution under the same conditions. Permeabilization of the spheroids was conducted at 4℃ for 2 days. The primary antibody was incubated for 2 days at 4℃. Next, the spheroids were washed 5 times in DW and then incubated with fluorescence-conjugated secondary antibodies for 2 days (If an antibody with fluorescence was used, the corresponding step was excluded). After washing 5 times with DW, nuclear staining was performed with DAPI at RT for 1 hour. Finally, the 96-well plate with spheroids was imaged and evaluated using fluorescence.

Cleared cardio-spheroid model for toxicity evaluation. (A) Optimized spheroid clearing protocol for the evaluation of cardiotoxicity. (B) Clearing improved the spheroid imaging results. Non-cleared cardio-spheroids (upper panel); Cleared cardio-spheroids (lower panel) were stained with DAPI (blue), α-actinin (red), and imaged by fluorescence microscopy; scale bar, 100 µm. (C) Fluorescence stacked images of three-dimensional spheroids stained with cTnT (green) and pro-B type natriuretic peptide (BNP, yellow) using a light-sheet microscope; scale bar, 50 µm.
We generated an hPSC-derived cardio-spheroid to verify the optimized spheroid clearing conditions and comparatively evaluated the fluorescence intensity after clearing through staining with the CM marker α-actinin. Nuclear staining was not significantly affected by clearing. In contrast, the antibody-stained spheroids showed increased image resolution and fluorescence intensity after clearing (Fig. 4B).
Next, the cardio-spheroids were treated with cardiotoxic drugs (levofloxacin and moxifloxacin) to validate whether they could be used as a model for drug toxicity evaluation after spheroid clearing. Moxifloxacin has been reported as a leading cardiotoxicity-inducing drug that prolongs the QT interval and increases the risk of ventricular arrhythmias [24]. In contrast, levofloxacin is a low-risk drug that is not associated with severe arrhythmias [25]. The cardio-spheroids were treated with 10 µM levofloxacin or 20 µM moxifloxacin. After 12 hours of incubation, the spheroids were fixed and cleared under optimized clearing conditions. The cleared cardio-spheroids were stained with pro–B type natriuretic peptide (BNP) and CM marker protein cTnT. pro-BNP is an important biomarker with an established role in the diagnosis of congestive heart failure. Furthermore, BNP has been demonstrated as a tool for the early prediction of anthracycline-induced cardiotoxicity [26]. We used pro-BNP as a marker of cardiotoxicity. Cleared spheroid imaging revealed that the expression of pro-BNP was higher in the spheroids treated with moxifloxacin, and the toxicity of levofloxacin was lower (Fig. 4C). It was verified that 3D bio-imaging of the entire cardio-spheroids and image-based toxicity prediction are possible through clearing.
Taken together, we established a spheroid clearing protocol and utilized it for image-based cardiotoxicity evaluation. Our clearing method enhanced the fluorescence image intensity of spheroids and showed whole spheroids. In addition, it was confirmed that our spheroid clearing technique can be applied for cardiotoxicity evaluation in a hPSC-based myocardial model.
Discussion
In the new drug development process, the focus on developing and platforming cell-based assays for more complex, biologically significant, and predictable drug screening has been accompanied by active research on 3D analysis models and related technologies [22-24]. Spheroids are similar to the in vivo environment and can be used as a favorable model for drug screening, as they have advantages in terms of reproducing the interactions between cells and substrates, including physiologically significant aspects [27,28]. Toxicity prediction and quantitative analyzes using spheroids are attractive as a research tool, but spheroid models are difficult to use for a wide range of screening applications because the experimental process of imaging and analysis is not straightforward. Moreover, image-based research is difficult to use in toxicity prediction models because the interpretation of results may differ depending on the skill level of the researcher or image resolution. To overcome this problem, we applied clearing technology to spheroids to improve the overall image resolution and build an easy-to-use platform for image-based toxicity prediction. The protocol we established increased the fluorescence sensitivity of antibody expression on the cell surface through clearing of the spheroids and made it possible to obtain overall images of the spheroids.
Organoids are 3D multicellular microtissues designed to closely mimic the complex structure and functionality of human organs that generally exhibit better in vivo cellular responses and interactions than 2D cell culture systems [29-31]. Accordingly, many studies have been conducted to produce organoids using stem cells, and human development and disease models have been reported for various tissues such as the small intestine, liver, and lungs [30,32-34]. The heart is a complex organ, and except for the recently reported cardioids [35], there have been no reports on the manufacture and implementation of cardiac organoids. Therefore, in this study, a 3D spheroid model that can reflect in vivo cell responses and interactions better than 2D cell culture was constructed and used in this experiment. Although the cardio-spheroids applied in this paper do not perfectly reflect heart tissue, it is suggested that they more closely resemble in vivo conditions and are somewhat closer to a heart organoid model than 2D cell culture conditions. In future work, by applying our clearing protocol to the recently published cardioid (cardiac organoid) model [35] for toxicity evaluation, a more advanced toxicity evaluation system can be established.
In this study, we established spheroid clearing techniques for cardiotoxicity assessment. In particular, it was possible to perform spheroid clearing, improve the image resolution, and obtain 3D bio-images of the entire cell tissue by using a light-sheet fluorescence microscope or high-throughput system images. The toxicity evaluation method using a high-throughput system can be used to analyze a large amount of samples quickly [36,37] and improve the accuracy of evaluation by incorporating clearing. Through this method, it is possible to obtain much faster and more precise results than with existing methods in disease model analysis using spheroids, and this approach can be used to identify the causes of disease and develop treatments.
Depending on the cell type and origin, questions may arise as to whether the protocol presented in this article can be applied to all organoid systems. Although the clearing method might need to be slightly modified according to cell origin, our protocol can be applied to HEK-293 spheroids, hPSC-derived cardio-spheroids, and hPSC-derived spheroids (data not shown), as well as commercially available hPSC-derived cardio-spheroids (data not shown). Based on these results, we suggest that the clearing of spheroids of different cell origins should be investigated through the application of our protocol.
It is clear that the clearing technique is a good system for toxicity evaluation, but a limitation of the method used in our study is that it is time-consuming. To overcome the limitations of the study, additional research should focus on performing toxicity evaluation more accurately and quickly by establishing conditions that reduce the time required for clearing. If more efficient toxicity evaluation is made possible by reducing the time required for clearing, the cardiac-spheroid clearing presented in this study will enable rapid and high-efficiency toxicity prediction and evaluation at the drug development stage. Furthermore, it is expected that patient-specific drug toxicity evaluation using iPSCs will be possible.
Notes
Conflict of interest
No potential conflict of interest relevant to this article was reported.
Funding
This research was funded by the National Research Foundation (NRF–2021R1F1A1061739) and the Korea Institute of Toxicology, Republic of Korea (1711133846).
Author contributions
Financial support: JHP, KKS; Investigation: JHP; Methodology: JML, SHP; Data analysis: JHP; Advice for experiment: JML, SHP, Writing-original draft: JHP; Final approval of the manuscript: KKS
Data availability
The dataset files are available from Harvard Dataverse at https://doi.org/10.7910/DVN/DNPZ8Z.
Data availability
Please contact the corresponding author for data availability.
