The application of intestinal organoids and their co-culture systems in the study of gastrointestinal diseases
Article information
Abstract
The 3-dimensional culture of intestinal organoids provides insights into the phenotype and physiology of the intestine. Intestinal organoids comprise intestinal stem cells and differentiated intestinal epithelial cells; they can be stably cultured over the long term and can be genetically manipulated. Current strategies for intestinal organoid co-culture with other cells existing in the intestinal tract and gut microbiota have been established to mimic the intestinal microenvironment and study host-microbial interactions. Therefore, intestinal organoids are promising tools for basic and translational research in gastroenterology. Gastrointestinal diseases are disorders of the intestinal tract that result in a reduced quality of life, and a deep understanding of these diseases would be effective for their treatment. In this review, we discuss how intestinal organoids and intestinal organoids integrated with cellular and microbiota niche components are biologically and physiologically relevant tools for the investigation of gastrointestinal diseases.
Introduction
Two-dimensional (2D) cell culture models and animal models were major tools widely used in biomedical research. Although 2D cell culture is a simple and low-cost method for maintaining cell culture, it also has many limitations due to the inaccurate representation of the complexity of in vivo conditions. Alternatively, animal models are useful in basic and translational research because they can recapitulate the nature of biologically relevant phenomena and pathologies observed in humans; however, they are time-consuming and costly. A new and improved method, the 3-dimensional (3D) cell culture technique, referred to as “organoids,” has been found to replicate much of the complexity of organs for clinical research. An organoid is a mini-organ formed in vitro in a 3D culture generated from stem cells or a group of cells. Organoids correspond to the phenotype and physiology that each organ possesses, such as self-organization, self-renewal, and differentiation to specific cell types. Therefore, organoid culture has the potential to provide alternative approaches to cover the gap between 2D cell culture systems and animal models. Various types of organoids have been developed to simulate the brain [1,2], liver [3,4], pancreas [4,5], kidney [6,7], stomach [8,9], and small intestine [10]. Moreover, organoids can be genetically manipulated to mimic disease-relevant genetic backgrounds, which is useful for mechanistic studies [11]. Nevertheless, the use of organoids for research is still subject to some limitations; for example, special techniques are required and there are complications related to assays.
The gastrointestinal (GI) tract is the most complex organ in the human body. It consists of multiple compartments, including the intestinal epithelium, gut microbiota, and the local immune system. The intestinal epithelium comprises diverse cell types organized along the crypt-villus axis and undergoes a renewal process to maintain homeostasis. Intestinal stem cells (ISCs) undergo asymmetric cell division into new stem cells and committed daughter cells, called transit-amplifying (TA) cells. TA cells subsequently differentiate into functional cell types, including absorptive enterocytes and secretory cells (goblet cells, Paneth cells, and enteroendocrine cells) [12]. The activity of ISCs can also be influenced by several factors, such as nutritional status and inflammation [13].
Intestinal organoids were first established from the culture of small intestinal crypts or single ISCs (adult stem cells) in a complex extracellular matrix (ECM) by Sato et al. [10]. Moreover, intestinal organoids were later generated from inducible pluripotent stem cells (iPSCs) or the human embryonic stem cell line H1 [14,15]. Intestinal organoids derived from adult stem cells and iPSCs have advantages and limitations; therefore, the selection of an intestinal organoid model depends on the purpose of the study [15]. For example, intestinal organoids derived from iPSCs may be better when studying the differentiation process of in vivo intestinal development, while intestinal organoids derived from adult stem cells may be useful for disease observation since the characteristics of the originating tissue will be preserved in organoids. Under physiological conditions, the crosstalk between intestinal epithelial cells (IECs) and other cell types in the GI tract plays a critical role in maintaining tissue homeostasis and promoting IEC survival and function. Therefore, to reflect the cellular heterogeneity and physiological relevance of intestinal tissue, a combination of intestinal organoid culture with stromal cells and immune cells was recently generated [16,17]. The gut microbiota and microbial metabolites are known to play an important role in the regulation of host health and disease, which can be mediated by the intestinal epithelium [18,19]. Likewise, the co-culture of intestinal organoids with gut microbiota has been recently reported [20]. These co-culture systems have improved researchers’ understanding of communication between IECs and the microenvironment. Intestinal organoid culture is useful in the study of GI diseases such as research aiming to establish disease models and drug discovery [21]. In this review, we highlight recent findings regarding the application of intestinal organoids and intestinal organoids integrated with cellular and microbiota niche components in the study of GI diseases.
Ethics statement: This study was a literature review of previously published studies and was therefore exempt from institutional review board approval.
Intestinal organoids as a tool for understanding GI biology
Intestinal organoids were initially generated from mouse small intestinal crypts containing Lgr5 ISCs [10]. Intestinal organoids comprise ISCs, TA cells, absorptive enterocytes, and secretory cells which maintain the basic crypt-villus structure as the original intestinal epithelial feature. Intestinal crypts were grown in a complex ECM (Matrigel) and growth medium supplemented with essential factors for proliferation and differentiation, including epithelial growth factor (EGF), noggin, and R-spondin, termed ENR [10]. The sorted Lgr5+ ISCs could also form intestines with features indistinguishable from those derived from whole crypts. The components of the growth media have been optimized to support the growth and characteristics of intestinal organoids. EGF is produced by mesenchymal stromal cells and Paneth cells, which can maintain ISC proliferation and are necessary for long-term culture [22]. R-spondin is a ligand of LGR5 and is mainly secreted by subepithelial fibroblasts. R-spondin plays a role in the maintenance of ISCs and increases intestinal organoid size and survival [23,24]. Noggin is a bone morphogenetic protein (BMP) antagonist that is mainly expressed by mesenchymal cells beneath the crypt [25]. BMP signaling limits the expansion of ISCs by suppressing the ISC signature genes [26]. Additional growth factors are required for the expansion of human intestinal organoids, such as the Wnt ligand Wnt-3A, a p38 inhibitor, and a transforming growth factor-β (TGF-β) inhibitor [27,28]. In addition, neuregulin 1 (NRG1), a key EGF family ligand produced by mesenchymal stromal cells, macrophages, and Paneth cells, is important for the proliferation of intestinal crypts and organoid formation, as well as for regeneration after injury [29]. A recent study also demonstrated that R-spondin 1 could be substituted by a compound named RS-246204 for mouse intestinal organoid culture [30]. Moreover, the modulation of Wnt and Notch signaling in intestinal organoids can regulate IEC lineage differentiation [31]. For example, the combination of inhibitor of Wnt production-2 (IWP-2) and valproic acid (VPA) could specifically induce enterocyte differentiation. The combination of the Notch inhibitor DAPT with other molecules, such as a glycogen synthase kinase 3 beta (GSK3β) inhibitor and IWP-2, could induce Paneth cell and goblet cell differentiation, respectively [31]. The modulation of IEC lineage differentiation in intestinal organoids is useful for studying specific IECs in response to stimuli or intestinal diseases.
In light of its unique properties in mimicking the in vivo environment, intestinal organoid culture is widely used for understanding GI biology and biomedical research [21]. Researchers have studied the responses of intestinal organoids to dietary nutrients, as well as gut microbiota and their microbial metabolites, which are commonly localized in the intestinal lumen [32,33]. Lee et al. [33] demonstrated that microbial-derived lactate can promote Lgr5+ ISC proliferation and intestinal epithelial regeneration as well as increase in organoid size and the number of Lgr5+ ISCs. The regeneration of small intestinal organoids after methotrexate or radiation-induced damage was observed when the organoids were treated with lactate. We utilized mouse small intestinal organoids to investigate the effect of vitamin D3 on the viability, stemness, and differentiation of IECs [32]. Vitamin D3 suppresses stemness and promotes apoptosis, while also inducing IEC differentiation into absorptive and secretory cells. Intestinal organoids have been used to generate models of intestinal disorders and disease development [34,35]. Hahn et al. [34] established an organoid-based epithelial-to-mesenchymal transition model for use in the study of intestinal fibrosis. Treatment with tumor necrosis factor-α (TNF-α) and TGF-β synergistically promoted the epithelial-to-mesenchymal transition in intestinal organoids. In addition, the culture of patient-derived intestinal organoids is now becoming a popular technique for understanding molecular mechanisms and developing methods for intestinal reconstruction [36,37]. Moreover, the IEC lineage can be modulated to mimic the characteristics of IECs, which is useful for studying the function of IECs under certain conditions. Overall, the application of intestinal organoids is very practical and useful for understanding GI biology under homeostasis or disease conditions.
Intestinal organoids integrated with cellular and microbiota niche components
The GI tract comprises multiple components, including the gut microbiota, intestinal stromal cells, and local immune cells, which affect the characteristics and functions of IECs. The intestinal lumen contains a huge community of gut microbiota, and crosstalk between the gut microbiota and IECs has been reported [38,39]. IECs express various types of receptors for the recognition of luminal gut microbiota and microbial metabolites [40,41]. The intestinal lamina propria contains stromal and immune cells. Intestinal stromal cells, including fibroblasts, myofibroblasts, pericytes, endothelial cells, and smooth muscle cells, are located in the mesenchymal elements surrounding the base of crypts [42]. Intestinal stromal cells can regulate IEC stemness, proliferation, and differentiation under conditions of homeostasis and damage [43,44]. Intestinal immune cells reside in the intestinal epithelium and lamina propria, and the interaction between intestinal immune cells and IECs promotes IEC differentiation and function [45,46]. Intestinal organoid co-culture with the gut microbiota, stromal cells, and immune cells has been established [17,20,47]. In this section, we discuss recent studies using intestinal organoids integrated with cellular and microbiota niche components in GI research. The models of intestinal organoids co-cultured with cellular and microbiota niche components are summarized in Fig. 1.

Intestinal organoids derived from different origins (iPSCs or adult stem cells) and the co-culture of intestinal organoids with other cellular components and the gut microbiota. iPSCs, inducible pluripotent stem cells.
1. Intestinal organoid co-cultures with the gut microbiota
The gut microbiota develops with the host, and different sections of the GI tract contain distinct populations of the gut microbiota [48]. The gut microbiota can utilize and convert ingested food and host products into microbial metabolites, which further impact host cells, including IECs [18]. Animal studies have shown that the development and function of the gut epithelium are abnormal in germ-free mice [49,50]. In contrast, the administration of enteric pathogens results in physiological dysfunction and inflammatory responses in IECs [51]. Studying gut microbiota and IEC interaction in vivo is highly complex, and in vivo research cannot demonstrate the direct interactions between these components. The use of cell lines to study these interactions cannot mimic the complex structure and function of IECs. Therefore, intestinal organoid culture has become an alternative tool with which to study the interactions between the gut microbiota and IECs. In addition, intestinal organoids derived from adult stem cells originate from the host tissue, which can provide the key features of mature IECs and cellular diversity mimicking the tissue of origin. Therefore, intestinal organoids derived from adult stem cells are more relevant and convenient than iPSCs.
Several techniques have been applied for the co-culture of intestinal organoids and gut microbiota, as shown in Fig. 2. The first technique is the fragmentation of intestinal organoids and replication of the gut microbiota. To this end, intestinal organoids were mechanically disrupted into small fragments and then re-plated with bacteria or bacterial products. Once the fragments were reassembled into organoids, the introduced gut microbiota or bacterial products were entrapped within the lumen. This model allows the bacteria to encounter the apical and basolateral sides of the IECs [52]. The infection of intestinal organoids with Listeria monocytogenes by the fragmentation method was used for proteomic analysis [52]. Muramyl dipeptide, a component of the bacterial cell wall, was introduced into intestinal organoids via fragmentation to study the effect of this bacterial molecule on IECs [53]. There are several limitations of this technique. For example, the number of bacteria entrapped in the lumen cannot be ensured, and the bacteria may randomly interact with the basolateral sides of IECs. The second technique is the microinjection of bacteria into the lumen of intact organoids [20]. This technique allows bacteria to come into direct contact with the apical side of IECs and is suitable for the investigation of single bacterial species’ effects on IECs. Several studies have been conducted to optimize this technique, including the calculation of injection volumes, the repeated microinjection method, and the use of fluorescent bacteria to assess the injection quality [54,55]. Yokoi et al. [56] showed that Paneth cells in intestinal organoids released granules in response to bacteria or lipopolysaccharide (LPS) when the organoids were exposed on the apical side by microinjection. However, this response did not occur when the organoids were exposed to LPS delivered in the culture medium to the basolateral surface. Taken together, microinjection is a powerful technique for studying the effect of gut microbiota or pathogens on IECs, particularly through interactions with the apical side. The advantage of this technique is that a precise dose of bacteria can be injected, and the hypoxic conditions of the lumen provide a suitable environment for the bacteria, which results in a longer experimental time for observation.
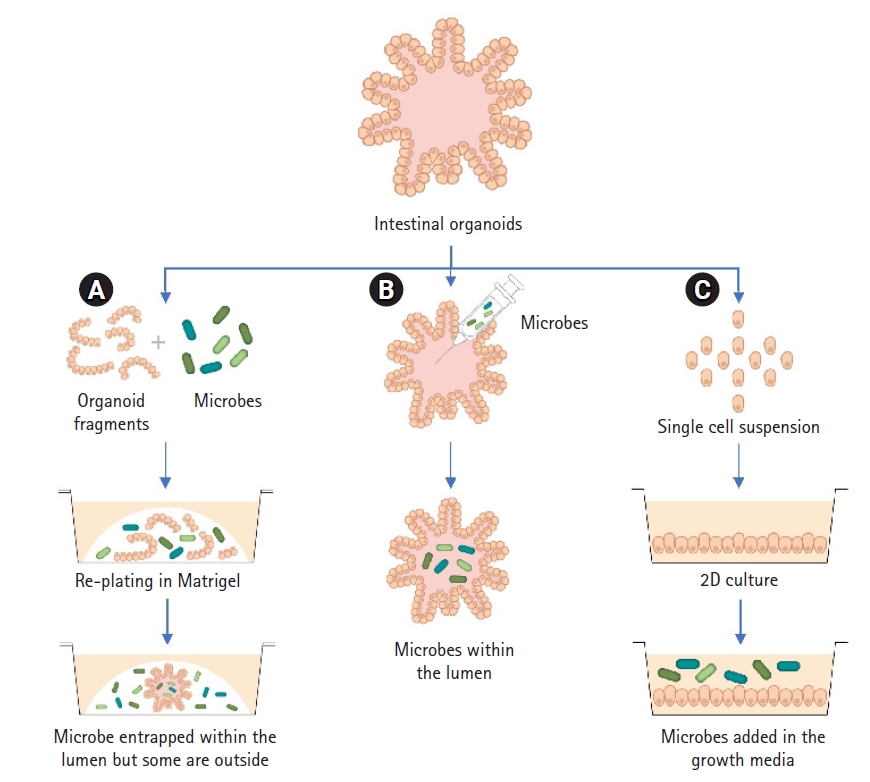
Methods of intestinal organoid co-culture with gut microbiota. (A) Organoids are sheared into smaller pieces, mixed with gut microbiota and re-plated with Matrigel. The gut microbes are randomly trapped within the organoid lumen after plating. (B) Introduction of bacteria into the intestinal organoids via the microinjection of bacteria into the lumen of intact organoids. (C) Organoids are dissociated into single cells and grown as a 2-dimensional monolayer. The gut microbiota are introduced into the medium.
There are limitations of microinjection technique that need to be addressed. For instance, it is difficult to scale up experiments because of the manual nature of the microinjection procedure. Microinjection is based on a single microbial species rather than a combination of bacteria, which may not reflect the complex community of the gut microbiota. A specialist is required to perform microinjections to minimize injection errors and organoid damage. Moreover, the high cost of the microinjection device may not be practical for certain studies. The modulation of organoid culture by controlling epithelial polarity may provide access to the apical side of IECs for gut microbiota to the apical side of IECs without disrupting the microinjection technique [57]. Alternatively, intestinal organoids can be dissociated into single cells and grown as 2D monolayers. This model provides the simplest approach with which to study interactions between the gut microbiota and IECs. 2D organoid-derived monolayers have been used to evaluate IEC function, such as permeability, cell differentiation, and the secretion of immune mediators in response to pathogens [58–60]. Cellular stress and reduced differentiation into secretory cells could be observed in the long-term culture of organoids as monolayers due to the generation of low oxygen tension. However, air-liquid interface culture allows the apical side of the monolayer to be exposed to the air, leading to the reduction of oxygen stress in the culture [61]. These techniques provide powerful tools for the study of gut microbiota-IEC interactions.
2. Intestinal organoid co-cultures with stromal cells
Intestinal stromal cells, such as fibroblasts and myofibroblasts, are located close to the crypt stem cells in the lamina propria [42]. Intestinal stromal cells produce mediators that are important for IEC functionality, such as stemness, proliferation, and differentiation [43,44,62]. Therefore, intestinal organoids need to be co-cultured with stromal cells to understand their coordinated biological functions. Moussa et al. [62] showed that the co-culture of intestinal organoids with mesenchymal stromal cells increased the number, proliferation, and size of intestinal organoids, which may be mediated by BMP antagonists. In addition, transplantation by the co-injection of intestinal organoids with mesenchymal stromal cells could yield improved therapeutic benefits for radiation-induced injuries [62]. Intestinal subepithelial myofibroblasts (ISEMFs) secrete growth factors, such as BMP antagonists and Wnt ligands, which provide a crucial signal for regulating ISC behavior [63]. The co-culture of intestinal organoids derived from adult stem cells with ISEMFs enables the long-term culture of intestinal organoids without the need for additional growth factors [64]. Growth and organoid formation were also enhanced when the intestinal organoids were co-cultured with ISEMFs [65]. Collagen-based culture systems are needed to compensate for the fact that Matrigel is not approved for clinical use. Jabaji et al. [66] demonstrated that intestinal organoids co-cultured with ISEMFs in collagen could be used for in vivo engraftment. Collagen-based methods cannot be used for the monoculture of intestinal organoids due to autolysis within 2 to 3 days. However, co-culture with ISEMFs or the addition of conditioned media from ISEMF culture was found to maintain the growth and structure of organoids in collagen [67]. Moreover, crypt-fibroblast co-culture in organotypic cell culture system could increase the organoid diameter and viability [16]. Taken together, these studies suggest that the growth factors produced by stromal cells in the co-culture of intestinal organoids with stromal cells are crucial for the survival and function of intestinal organoids.
3. Intestinal organoid co-cultures with immune cells
Several subsets of innate and adaptive immune cells are present in the intestinal epithelium and intestinal lamina propria, and they can modulate intestinal homeostasis [68]. Lamina propria immune cells can recognize and respond to the gut microbiota or bacterial metabolites, which in turn influences IEC functionality. For example, immune cells and immune mediators or cytokines can modulate the self-renewal and differentiation of ISCs [69]. Multiple approaches can be used for immune cell and/or cytokine co-culture with intestinal organoids. First, the treatment of intestinal organoids with cytokines is used to estimate the effect of immune cell-derived cytokines on IECs. Second, to demonstrate the interactions between immune cells and IECs. For example, Noel et al. [70] co-cultured primary human macrophages with intestinal organoids using 2D organoid-derived monolayers to investigate intestinal barrier function and host-pathogen interactions. They found that macrophages could enhance the barrier function and maturation of IECs. In addition, macrophages extend dendrites through the organoid monolayer and interact with Escherichia coli in the lumen-mimicking compartment, resulting in a cellular response to pathogens. Biton et al. [69] found that the number of TA cells was higher in the intestinal organoids co-cultured with polarized T helper (Th) cells or treated with interleukin (IL)-17. In addition, co-culture with regulatory T cells or treatment with IL-10 promoted ISC expansion in intestinal organoids. Moreover, the expression of major histocompatibility complex class II, an antigen-presenting molecule, in Lgr5+ ISCs was induced by Th1 cell molecules, which may initiate immune cell-IEC crosstalk. The growth of mouse intestinal organoids was increased in organoids co-cultured with innate lymphoid cells, a potent IL-22 producer, compared with monocultured organoids [71]. Moreover, IL-22 treated organoids showed increased proliferation and ISC expansion. Intestinal organoids derived from iPSCs co-cultured with T cells and treated with IL-2 resulted in the maturation of organoids that exhibited the characteristics of mature adult intestinal epithelium [72]. Together, these findings suggest that co-culture models of intestinal organoids with immune cells or cytokines are powerful tools for the study of the bidirectional communication between immune cells and IECs, as well as the response of IECs to immune cells or cytokines and vice versa.
Intestinal organoids in the study of GI disease
Intestinal organoid culture has been increasingly developed, proving to be useful for pathological studies and translational applications, such as disease modeling, drug screening, and regenerative medicine. Several studies have utilized intestinal organoid culture as a model for several GI disorders, such as GI infectious diseases [73,74] and inflammatory bowel disease (IBD) [75,76]. Studies using intestinal organoid culture to model GI disorders are discussed in this section.
1. GI infectious diseases
Intestinal organoids have been used to study the interactions between the host and pathogen, as well as common infectious agents. There are several approaches to the study of infectious diseases using intestinal organoids. Intestinal organoids can be exposed to pathogens or infectious agents using 3 different methods [77]. First, intestinal organoids can be exposed to pathogens by microinjection, which enables direct contact between the apical side of IECs and pathogens. Second, organoids can be mechanically dissociated into smaller aggregates and incubated with pathogens before re-seeding into Matrigel. Third, organoids can be enzymatically digested into single cells and grown as 2D organoid-derived monolayers, whereafter the pathogen is added to the culture media. Infection with Clostridium difficile is a common cause of hospital-acquired diarrhea, resulting in increased healthcare costs and high mortality [78]. The pathogenicity of this pathogen is mediated by C. difficile toxin A (TcdA) and C. difficile toxin B (TcdB), which cause fluid secretion, inflammation, and intestinal damage [79]. Intestinal organoids have been extensively used to elucidate the pathology of C. difficile in IECs [73,74,80]. Mature human intestinal organoids have been used to assess C. difficile toxin function and intestinal damage [80]. Mileto et al. [73] investigated the direct effects of ISC infection, and found that the function and regeneration capacity of ISCs were compromised in intestinal organoids derived from C. difficile-infected mice, with these effects being mediated by TcdB. The microinjection of C. difficile or purified TcdA into human intestinal organoids induced the disruption of epithelial paracellular barrier function [74]. TcdB is known to inhibit the activity of the NHE3 sodium/hydrogen exchanger in IECs [81]. To confirm and extend these findings, Engevik et al. [82] showed that the expression of NHE3 was decreased in human intestinal organoids microinjected with C. difficile or stool supernatant of C. difficile-infected patients. Salmonella, an invasive enteric pathogen, can interact with and pass through the intestinal mucosa, resulting in gastroenteritis [83]. Zhang et al. [84] showed that bacterial adherence and invasion of IECs were observed after the organoids were colonized with Salmonella for a short period. Furthermore, Salmonella infection caused the onset of an inflammatory cascade mediated by NF-κB activation and a reduction of ISC markers. RNA sequencing analysis showed an altered transcriptional signature, including the patterns of cytokine expression, in human organoids derived from iPSCs microinjected with Salmonella. In addition, Salmonella microinjected into the lumen of intestinal organoids exhibited invasion of the infected organoids, which mimics the ability of Salmonella to invade IECs [85]. Taken together, these studies provide an excellent model for the study of GI infectious diseases, especially the response of IECs to pathogens or infectious agents, as well as the characteristics of pathogens in the interaction with IECs.
2. IBD
IBD is one of the most common intestinal diseases, with an increasing incidence worldwide. It is subdivided into Crohn’s disease and ulcerative colitis (UC), which is characterized by repetitive inflammation in the GI tract [86]. An inappropriate immune response causes intestinal barrier dysfunction, which plays a role in the pathogenesis of IBD. Intestinal organoid culture has been used to study epithelial barrier function in IBD [76,87]. Xu et al. [87] established a model using intestinal organoids derived from patients with IBD. Intestinal integrity was observed by the permeation of fluorescein isothiocyanate-labeled dextran 4 kDa (FITC-D4), into the lumen of organoids and the expression of key tight junction genes. Intestinal organoids derived from patients with IBD grew normally and preserved the transcriptional signature of IECs observed in patients [88]. The expression of a transcription factor controlling differentiation into the secretory lineage was decreased in differentiated organoids derived from patients with IBD [89]. Recently, induced human UC-derived organoids (iHUCOs) have been established [75]. Colonic fibroblasts isolated from patients with UC were re-programmed into iPSCs, followed by directed differentiation to iHUCOs. The developed iHUCOs had both epithelial and stromal compartments, as well as histological and functional features of primary tissue, such as impaired adherent junctions, aberrant proliferation, and a lack of goblet cells. In a mouse model of chemically induced colitis, dextran sodium sulfate (DSS)-induced colitis is regularly used to mimic the pathogenicity of human UC [90]. Although the mode of action of DSS remains unknown, it causes damage to the epithelial barrier and increases intestinal permeability for luminal bacterial components. Rallabandi et al. [76] developed an inflammatory IBD intestinal organoid model by DSS induction to study morphology and permeability. DSS-treated organoids showed damage to the epithelial barrier and increased inflammatory signaling-related genes involved in IBD, such as IL-6 and TNF-α, suggesting that DSS-treated organoids have a physical functionality, as is commonly observed in in vivo models. IL-22 is a critical regulator of intestinal barrier protection by regulating permeability, as well as promoting tissue regeneration–linked antimicrobial peptide responses in the intestinal mucosa [91]. To confirm and extend these findings, intestinal organoid cultures have recently been used [92,93]. Human intestinal organoids and the co-culture of organoids with primary human T cells were used to study the effect of IL-22 on ISC expansion and cell proliferation [92]. This study showed that IL-22 enhanced the growth and number of Ki67-positive cells in organoids in a concentration-dependent manner. In addition, IL-22 also promoted antimicrobial response in either a single culture of intestinal organoids or co-culture of organoids with immune cells. Moreover, Aden et al. [93] evaluated the interplay between IL-22 and the IBD risk gene ATG16L1 using mouse intestinal organoids. Taken together, intestinal organoids provide unique insights into individual patient status and constitute a potential method for the development of personalized treatment.
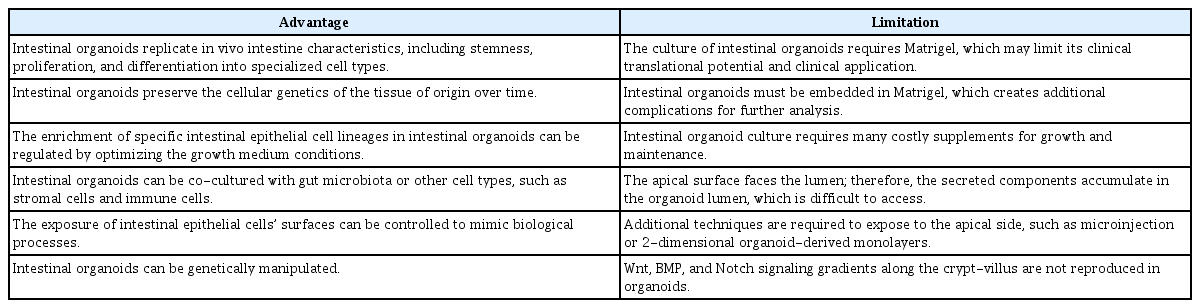
Advantages and limitations of intestinal organoids
Intestinal organoids are novel research tools for studying intestinal function and pathology. However, several limitations associated with the use of intestinal organoids remain. The advantages and limitations of using intestinal organoids are addressed in this section, and are summarized in Table 1.
1. Advantages
Intestinal organoids replicate in vivo intestinal characteristics, including stemness, proliferation, and differentiation into specialized cell types. Intestinal organoids can be cultured over the long term while maintaining their original properties [10]. In addition, intestinal organoids preserve the cellular genetics of the tissue of origin over time. Intestinal organoids can be derived from either adult stem cells or iPSCs, depending on the purpose of the study. For example, intestinal organoids derived from adult stem cells preserve the original characteristics of the tissue of origin, which is useful when studying the functionality of IECs and intestinal disease biology. In contrast, intestinal organoids derived from iPSCs are useful for the study of intestinal development. The enrichment of specific IEC lineages in intestinal organoids could be regulated by optimizing the growth medium conditions. For example, the combination of the Notch inhibitor DAPT with a GSK3β inhibitor could promote Paneth cell differentiation, while the combination of IWP2 with VPA could induce enterocyte differentiation in intestinal organoids [31]. Intestinal organoids can be co-cultured with gut microbiota or other cell types, such as stromal cells and immune cells, as described above. The co-culture system is very useful for understanding the crosstalk between IECs and other elements in the GI tract. In addition, intestinal organoids respond to soluble stimuli, such as microbial-derived molecules, microbial metabolites, and cytokines, supplemented in the growth medium. The exposure of IECs to either apical or basolateral cell surfaces could be controlled to mimic biological processes observed in vivo. Conventional intestinal organoid culture allows the apical side of IECs to face the lumen. The microinjection technique is generally used to introduce the gut microbiota to the apical side, which is suitable for the direct interaction between gut microbiota and IECs [20,56]. Intestinal organoids can be genetically manipulated, which is important when studying the role of genes in intestinal function in normal physiology and diseases. Several techniques have been established to modulate genes of interest in organoids, such as lentiviral transduction [94], liposomal transfection [95], and electroporation of gene vectors [96]. In addition, the clustered regularly interspaced short palindromic repeats (CRISPR)/CRISPR-associated (Cas) system has been recently developed to accurately manipulate genomic sequences in organoids [97].
2. Limitations
Some limitations of intestinal organoid culture remain to be considered. Organoid culture requires an ECM, and Matrigel is commonly used. Matrigel, a heterogeneous and complex mixture of ECM proteins, has lot-to-lot compositional and structural variability. In addition, Matrigel may not contain the necessary signal components for organoid structure and function. Moreover, Matrigel originates from mouse cells, which can cause immunogenicity in humans after transplantation, limiting its clinical translational potential and clinical use [98]. Alternative materials, such as other naturally derived proteins and biomacromolecules, are needed to overcome this limitation [67,98,99]. For example, type I collagen has been used to form intestinal organoids originating from isolated Lgr5+ progenitor cells or intact crypts, which can be used for the regeneration of damaged mouse intestinal epithelia upon transplantation [100]. In addition, organoids must be embedded in Matrigel, which creates additional complications for further analysis. Organoids must be removed from Matrigel prior to processing or other procedures, such as RNA and DNA isolation, as well as genetic manipulation. Furthermore, intestinal organoid culture requires many supplements for growth and maintenance, which is costly. The apical surface of IECs faces the lumen and the basolateral surface faces the outside; therefore, the secreted components accumulate in the organoid lumen, which is difficult to access for the study of the secreted components in the lumen. Moreover, additional techniques are needed to expose the molecules of interest to the apical side of IECs in organoids, such as microinjection or 2D organoid-derived monolayers [20,58–60]. Wnt, BMP, and Notch signaling gradients along the crypt-villus, which are crucial for stemness, proliferation, and differentiation, are not reproduced in current intestinal organoid culture systems. In addition, scalability and reproducibility are limitations of intestinal organoid use. Therefore, extensive further study is required to overcome the limitations of the culture and application of intestinal organoids.
Conclusion and future perspectives
Intestinal organoids have been used as alternative tools for the study of intestinal function and pathology. Intestinal organoids can be integrated with cellular and microbiota niche components to improve biologically and physiologically relevant tools for intestinal research. However, several limitations of intestinal organoids remain to be addressed. For example, Wnt, BMP, and Notch signaling gradients along the crypt-villus are not reproduced in current intestinal organoid culture systems. The regulation of stemness, proliferation, and differentiation by these signaling gradients is important for maintaining tissue homeostasis and regeneration. Therefore, strategies to control the levels of these crucial signals are required. Intestinal organoids also have an apical surface facing the lumen and a basolateral surface facing the outside. As a result, the secreted components accumulate in the organoid lumen. However, at present, no methods for the sampling and assessment of secreted molecules in the organoid lumen exist. Thus, to expose the apical surface of IECs in organoids, a microinjection technique is required, which may damage the structure of organoids, as well as requiring expert skill when performing the experiment. Therefore, alternative strategies for exposing the apical surface are needed. Although current intestinal organoid culture systems have some limitations, organoid culture application has contributed to GI research. Intestinal organoid culture is being optimized as a better model for basic and translational research.
Notes
Conflict of interest
No potential conflict of interest relevant to this article was reported.
Funding
This work was supported by a grant from the National Research Foundation of Korea (2021R1I1A1A01045477) to PS and (2020R1I1A3073474, 2021M3A9I4027993, 2017M3A9F3043837) to YKL.
Data availability
Please contact the corresponding author for data availability.