Three-dimensional cardiac organoid formation accelerates the functional maturation of human induced pluripotent stem cell-derived cardiomyocytes
Article information
Abstract
Background
Human induced pluripotent stem cell (hiPSC)-derived cardiomyocytes (CMs) offer a promising source for heart disease modeling and drug screening. Recent developments in organoid technology have made it possible to study how hiPSC-derived CMs interact together, and this culture system mimics the tissue environment and behavior of the cardiac cells in our body. However, the similarities and differences between conventional 2-dimensional (2D) culture and 3-dimensional (3D) organoid culture systems for CM differentiation have been incompletely elucidated.
Methods
To study how the individual microenvironment formed by each culture system affects the properties of CMs differentiated from hiPSCs, we conducted a comparative study between 2D monolayer and direct 3D cardiac organoid differentiation from hiPSCs throughout the sequential differentiation stages.
Results
The 3D differentiation system strongly exhibited higher mesoderm commitment and cardiac induction than 2D monolayer differentiation from hiPSCs. In the late stage of differentiation, the 3D cardiac organoids showed a higher frequency of a mature myofibrillar isoform switching in sarcomere structure of differentiated CMs than was observed in monolayer culture, although over 94% of cardiac troponin T-positive cells resulted at the end point of differentiation in both systems. Furthermore, the accelerated structural maturation in 3D cardiac organoids resulted in increased expression of cardiac-specific ion channel genes and Ca2+ transient properties, with a high signal amplitude and rapid contractility.
Conclusion
The present study provides details surrounding the 2D and 3D culture methods for CM differentiation from hiPSCs and focuses on 3D cell culture as an improved strategy for approaching and applying cardiac maturation.
Introduction
Based on the enormous progress in stem cell research, the regulation of key signaling pathways in the developmental process of specific organs in our body enables the direct differentiation of human pluripotent stem cells (hPSCs), including human embryonic stem cells and human induced pluripotent stem cells (hiPSCs), into any desired cell type [1,2]. In particular, several well-defined protocols such as the sequential addition of small molecules and/or supplements (e.g., growth factors) in hPSC cultures have successfully guided the differentiation of selective cardiac cells such as cardiomyocytes (CMs), which have been considered an attractive in vitro model for various applications [3–6].
However, traditional differentiation systems based on 2-dimensional (2D) monolayer culture could not provide a sufficient environment to mimic human heart development and pathophysiology because cardiogenesis in the human embryo is a very delicate and complex process characterized by intricate spatiotemporal dynamics of soluble biochemical and biophysical stimuli [7,8]. Furthermore, the conventional cardiac differentiation on 2D monolayer culture bears a few disadvantages: First, the initial seeding density of undifferentiated cells results in fluctuations in the differentiation efficacy of CMs, leading to batch-to-batch variation in CM yield and purity [5,9–13]. Second, cardiac differentiation in 2D monolayer culture systems still requires a harsh purification step during the differentiation process to obtain a highly pure population of CMs, because unwanted cell types, including those belonging to the endodermal or ectodermal lineage, result in a low yield of cardiac differentiation [14–16]. However, this purification process, which includes glucose starvation, may also result in unhealthy cardiac differentiation.
Three-dimensional (3D) structures such as organoids or aggregates have emerged as powerful platforms for advanced differentiation, since the several cell types grown in the 3D microenvironment more closely resemble in vivo cells both morphologically and in their genetic/molecular regulation [17]. Therefore, in recent decades, various studies have reported the advantages of 3D in vitro models for many cell types [8]. Most studies have shown that 3D organoids can mimic in vivo organogenesis, including the behavior of cells in the organ, and these models have been applied in the field of stem cell research, research pertaining to human disease, and drug discovery. Along with the advantages of 3D organoid models for many cell types, the benefits of cardiac differentiation in 3D culture systems have also been reported [18–20]. As an example, Correia et al. [21] studied the benefits of a 3D platform at earlier stages of cardiac differentiation, including reinforcement of the maturity of structural and metabolic information in stem cell-derived CMs, such as internal cytoskeletal organization and integrity.
In the present study, we sought to advance the knowledge of hiPSC-derived CM differentiation according to the culture system used for differentiation by investigating the expression of selected genes related to heart development, cellular maturation, and cardiac-specific ion channels during cardiac differentiation of hiPSCs in 2D monolayer and 3D structures. We further examined real-time measurements of calcium flux in 2D CMs and 3D cardiac organoids, reflecting electrophysiological and Ca2+ signaling properties. Ultimately, this study focused on the characterization of an optimized 3D culture system for the derivation of hiPSCs into CMs to strengthen their function compared to the conventional 2D differentiation platform.
Materials and Methods
Ethics statement: The fibroblasts used for generating iPSCs in this study was purchased from commercially avialible vendor, ATCC, and was therefoe exempt form institutional review board approval.
1. Expansion and maintenance of hiPSCs
The hiPSCs were initially generated and characterized by NEXEL Co., Ltd. (Seoul, Korea), using Sendai virus transduction of BJ fibroblasts (ATCC Inc., Manassas, VA, USA) with 4 retroviral reprogramming factors. The established hiPSCs (46, XY) were expanded and maintained in feeder-free conditions in T75 tissue culture flasks coated with Matrigel at a 1:100 ratio in DMEM/F12 with mTeSR 1 medium (StemCell Technologies, Vancouver, Canada). The culture medium was changed every day. After 80-90% confluence was reached, hiPSCs were passaged using the TrypLE dissociation buffer (Invitrogen, Carlsbad, CA, USA). The cells were cultured at 37°C under 5% CO2.
2. Two-dimensional monolayer CM differentiation
The hiPSC-derived 2D cardiomyocytes (2D-CMs) used in this study were produced according to the standard differentiation protocol for the Cardiosight-S from NEXEL Co., Ltd. Briefly, Matrigel was diluted 1:100 in ice-cold DMEM/F12 to coat wells in the culture plate. The coated plates were incubated at 37℃ for at least 30 minutes, and after plating, the hiPSCs were seeded in Matrigel-coated wells using mTeSR 1 medium with 10 μM Y27632. The medium was changed every day. When the hiPSCs reached 80% confluence, they were treated with a combination of 10 μM activin A, 10 μM BMP4, and 4 μM GSK inhibitor for 2 days, followed by 5 μM Wnt/ꞵ-catenin inhibitor treatment for 2 additional days. On day 6, the medium was changed to RPMI 1640 medium with B-27 supplementation without vitamin A by changing the medium every 2 days. On day 11, the medium was changed to purification medium for metabolic high-purity CM differentiation, and the cells were maintained in RPMI 1640 medium with B-27 supplementation without vitamin A when the differentiation was finished.
3. Generation of cardiac organoids
For 3D cardiac organoid culture, we used the method of embryoid body (EB) formation and maintenance on ultra-low-attachment (ULA) 6-well plate (~2.0×106 cells per well) to allow 3D aggregate formation by undisturbed incubation for 4 days in mTeSR 1 medium with 10 μM Y27632. Cardiac differentiation was initiated approximately 4 days after seeding (D0) and performed through small-molecule control of the Wnt/ꞵ-catenin pathway with GSK3 inhibitor in RPMI 1640 supplemented with B-27 without insulin for 48 hours and inhibition of Wnt/ꞵ-catenin pathway for 48 hours. On day 6, the medium was changed to RPMI 1640 medium with B-27 supplementation without vitamin A by changing the medium every 2 days. The hiPSC-derived cardiac organoids (hiCOs) spontaneously began beating around day 8 of differentiation and they were maintained in RPMI 1640 medium with B-27 supplementation without vitamin A until day 24 (D24) for analysis.
4. Flow cytometry analysis
For the flow cytometry analysis, monolayer 2D-CMs were dissociated into single cells using a combination of 0.05% trypsin-EDTA with TrypLE for 20 minutes, and 3D hiCOs were dissociated into single cells using 0.05% trypsin-EDTA for 20 to 30 minutes at 37℃ in shaking incubators. Dissociated single cells were filtered through a 100-μm mesh cell strainer and simply treated with Fix/Perm Solution (BD Biosciences, San Jose, CA, USA) for 20 minutes at 4°C for one step of fixation and permeabilization. The cells were washed with 1× wash buffer (BD Bioscience) 3 times and incubated with primary cardiac troponin T (cTnT) antibody in a 1:200 dilution for 60 minutes at 4℃ in 1× wash buffer. Finally, the cells were incubated with Alexa Fluor 488 secondary antibody for 45 minutes at 4℃, and washed with 1× wash buffer 3 times. The cell suspension was analyzed on ACEA NovoCyte-3000 Flow cytometer (ASEA Bioscience, San Diego, CA, USA).
5. Quantitative real-time polymerase chain reaction
On day 24 of differentiation, total RNA from hiPSC-CMs was harvested using the traditional TRIzol method. The 3D hiCOs were prepared in TRIzol reagent (Thermo Fisher Scientific, Waltham, MA, USA) and were ultrasonicated for 4 minutes on ice using a sonicator (Sonoplus mini-20; 20 seconds on/off, ultrasonic on/off cycle: 0.5 minutes, wave output power: 320W). Immediately, RNA was isolated from the prepared samples. The RNA was quantified with a SpectraMax iD5 Microplate Reader (Molecular Devices). Total cDNA was synthesized by performing reverse-transcription polymerase chain reaction (PCR) using a ReverTra Ace quantitative PCR (qPCR) RT Master Mix with gDNA Remover (Toyobo, Osaka, Japan) according to the manufacturer’s instruction. For real-time qPCR, the cDNA was amplified and quantified using TB Green Premix Ex Taq (Tli RNaseH Plus) Kit (Takara, Shiga, Japan) on CFX Connect Real-Time PCR detection system (Bio-Rad, Hercules, CA, USA). The amplification reactions were as follows: 95℃ for 30 seconds, 44 cycles at 95℃ for 5 seconds, 60℃ for 30 seconds, dissociation at 55°C to 60℃ for 5 seconds depending on each primer, and 95℃ for 5 seconds. GAPDH was used as the housekeeping gene as an internal reaction control to normalize the relative quantification in each expression level, followed by calculations using the 2-∆∆CT protocol. Genes of interest included those involved in cardiac development, cardiac maturation, and CM-specific genes for cardiogenesis, and the primer sequences were designed using Primer3. The primer sequences used in this study are specified in Table 1.
6. Western blot analysis
On day 24 of differentiation, cell lysates of 2D-CMs were collected by scraping with radioimmunoprecipitation assay (RIPA) lysis buffer containing a protease and phosphatase inhibitor cocktail. The 3D hiCOs were prepared in RIPA lysis buffer and were ultrasonicated for 4 minutes on ice using a sonicator (Sonoplus mini-20; 20 s on/off, ultrasonic on/off cycle:0.5 minutes, wave output power: 320 W). The obtained protein samples were quantified by a bicinchoninic acid protein assay kit (Thermo Fisher Scientific) to load equal concentrations of protein. Next, 20 μg of protein was separated on a gradient of 4% to 12% Tris-glycine gels (Thermo Fisher Scientific) and electrophoretically transferred to 0.2 μM nitrocellulose transfer membranes on an iBlot 2 Dry Blotting System (Thermo Fisher Scientific). The membranes were blocked with 5% skim milk and probed with appropriate primary antibodies (1:1,000 to 1:10,000) overnight at 4℃. Then, the membranes were incubated with poly-clonal anti-rabbit/mouse horseradish peroxidase-conjugated secondary antibodies for 45 minutes at room temperature, and finally the membrane was developed using a Pierce ECL Western Blotting substrate.
7. Intracellular Ca2+ transient assay
The monolayer 2D-CMs used for intracellular calcium influx analysis were Cardiosight-S from NEXEL Co., Ltd., which were cultured according to the manufacturer’s protocol. First, 1×105 2D-CMs were seeded in fibronectin-coated (Sigma-Aldrich, St. Louis, MO, USA) wells of a clear flat-bottom black 96-well culture plate (final concentration of fibronectin: 50 μg/mL). For loading hiCOs on the plate, a 10 μM Matrigel droplet diluted in DMEM/F12 at a ratio of 1:2 was added in each well for 45 minutes. Matrigel maintains a dome shape during coating. After aspirating the Matrigel coating solution, a certain number of hiCOs suspended in 10 μL of RPMI 1640 were seeded as a droplet at the same spot where the Matrigel was coated, and they were centrifugated at 300×g for 3 minutes to form an organoid cluster. The seeded 2D-CMs and 3D hiCOs were maintained with RPMI 1640 for 7 days by replacing the medium every 2 days. On the day of the assay, the medium of 2D-CMs and 3D hiCOs was changed to DMEM/F12, including 1.8 mM calcium chloride (CaCl2) at least 3 hours prior to the test.
For calcium dye loading, the medium was changed by 90 μL of RPMI 1640 without supplements, after which 10 μL of FLIPR Calcium 6 dye (Molecular Devices, San Jose, CA, USA) solubilized in DMSO at 1:250 was added to each well of the plate. The 96-well plate was stored in the 37℃ incubator for 45-60 minutes before detection of intracellular Ca2+ influx. Spontaneous fluorescence readings were obtained on a SpectraMax iD3 multi-mode microplate readers (Molecular Devices) at 10 data points per second for 10 s. The systems have a heated stage that is maintained at 37℃ during recording for maintaining cardiac conduction. The Ca2+ imaging and the raw data were exported to Microsoft Excel software (or XML files), and then we analyzed the amplitude and beating rate (beats per minute) of individual 2D-CMs and 3D hiCOs via an in-lab-developed calculation method. For visualizing calcium influx, images were then acquired using a Nikon laser-scanning fluorescence microscope with a 20× objective (Nikon, Tokyo, Japan). Representative images were acquired for 10 s and quantitative imaging was analyzed using NIS-Elements AR software version 5.3 (Nikon) by the extent of green fluorescence (FLIPR Calcium 6 dye) intensity on live 2D-CMs and 3D hiCOs.
8. Contractility recordings on the FLEXcyte 96 system
The monolayer 2D-CMs used for contractility analysis were Cardiosight-S from NEXEL Co., Ltd.), which were cultured according to the manufacturer’s protocol. Briefly, 5 to 10×104 2D-CMs were seeded on a fibronectin-coated (Sigma-Aldrich) well on a 96-well plate with flexible silicon membranes from Nanion Technologies (Munich, Germany). For loading 3D hiCOs on the plate, a fibronectin droplet diluted in phosphate-buffered saline (Welgene, Daegu, Korea) was added to each well for at least 2.5 hours (final concentration of fibronectin: 50 μg/mL for 2D-CMs; 100 μg/mL for 3D hiCOs). Fibronectin maintained the dome shape during coating. After aspirating the fibronectin coating solution, a certain number of 3D hiCOs suspended in 20 μL of RPMI 1640 (Thermo Fisher Scientific) were seeded as a droplet at the same spot where the fibronectin was coated in each well. The seeded 2D-CMs and 3D hiCOs were maintained with RPMI 1640 for 7 days by replacing the pre-warm maintenance medium every 48 hours in a cell culture incubator at 37°C under 5% CO₂ conditions. Then, a day before the FLEXcyte 96 measurements, the medium in each well was replaced with a fresh pre-warm maintenance medium. The FLEXcyte 96 measurements were obtained using DataControl software (Nanion Technologies).
9. Statistical analysis
All experimental data were analyzed using GraphPad Prism 9 (GraphPad, San Diego, CA, USA) and Excel (Microsoft, Redmond, WA, USA) software for statistics and graph production. All data are presented as the mean±standard error of the mean, and the statistical significance of each result was determined using the Student t-test or one-way or 2-way analysis of variance with post hoc testing using the Dunnett method; p<0.05, p<0.01, and p<0.001 were considered to indicate statistical significance against 2D hiPSCs at each day. p<0.05, p<0.01, and p<0.001 were considered to indicate statistical significance against 2D hiPSCs on D0, D8, or D24.
Results
1. Three-dimensional cardiac differentiation enhances mesodermal commitment, cardiac specification, and differentiation of hiPSCs
To induce cardiac differentiation of hiPSCs, we adopted temporal modulation of the Wnt/ꞵ-catenin pathway for mesoderm commitment, and the standard protocol for inducing cardiac progenitor and maturation of cardiac cells was applied [22–24]. However, the cardiac progenitors from hiPSCs in the 2D monolayer differentiation system underwent the temporal metabolic purification phase during differentiation, whereas the differentiated 3D hiCOs from EB formation were cultured on the differentiation medium without a purification phase in ultra-low-attachment plates (Fig. 1A).
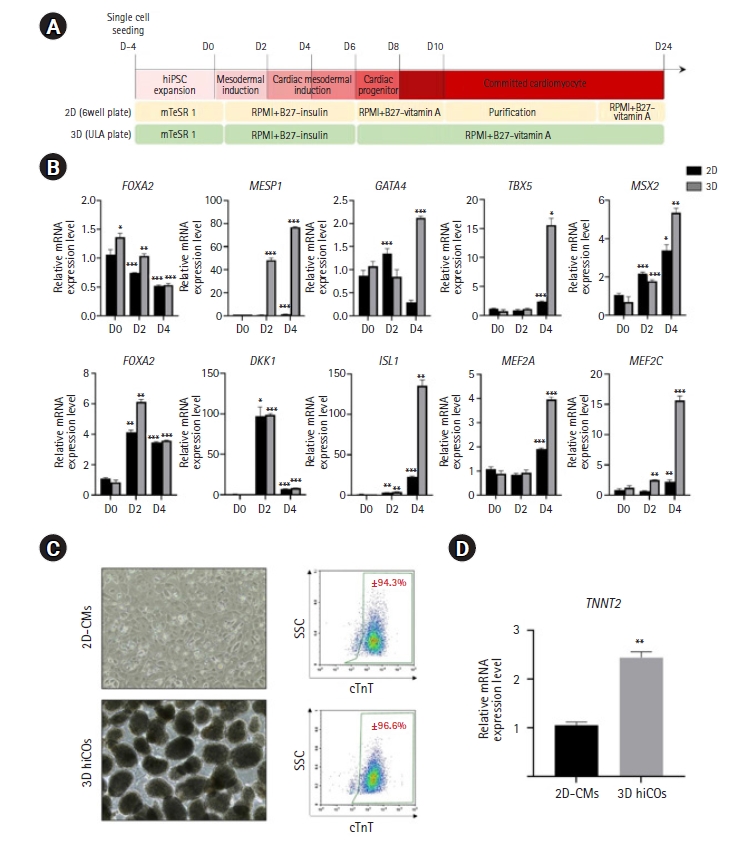
Human induced pluripotent stem cell-derived cardiac oragnoids (hiCOs) display high efficiency of the mesodermal subtype. (A) Schematic representation of 2-dimensional (2D) cardiomyocyte and 3-dimensional (3D) cardiac organoid protocols. (B) Real-time polymerase chain reaction analysis at the early stage of differentiation. Expression profile of the FOXA2, MESP1, GATA4, TBX5, MSX2 AXIN2, DKK1, ISL1, MEF2A, and MEF2C genes in the early stage (D0-D4) of differentiation. Values are normalized to GAPDH and relative to day 0. The statistical analyzes were conducted by 2-way ANOVA. (#p<0.05, ##p<0.01, and ###p<0.001 versus 2D human induced pluripotent stem cells [hiPSCs] at D0; n=3, *p<0.5, **p<0.01, and ***p<0.001 versus 2D hiPSCs on each day; n=3). (C) Representative optical images and cardiac troponin T (cTnT)-positive cells from 2D-cardiomyocytes (CMs) (×100 magnification) and 3D hiCOs (×40 magnification)analyzed by flow cytometry on D24. No apparent difference in differentiation efficiency was observed between 2D-CMs and 3D hiCOs. (D) mRNA expression levels of cTnT in 3D hiCOs (SSC indicates side scatter). Data are expressed as the mean±standard error of the mean (*p<0.5, **p<0.01, and ***p<0.001 versus 2D hiPSCs on each day; n=3). ULA, ultra-low-attachment.
A real-time PCR analysis from D0 to D4 differentiation revealed differences in the early lineage determinants for differentiating hiPSCs in 2D and 3D culture systems. FOXA2, a transcription factor known to be related to early/anterior primitive streak formation, presented peak expression at D0 and its expression gradually decreased up to D4 in both differentiation systems (Fig. 1B). In contrast, the expression levels of MESP1, GATA4, TBX5, and MSX2, transcription factors related to mesodermal and cardiac specification, gradually increased during the differentiation period from D0 to D4; however, the expression levels of differentiating cells in the 3D culture system were significantly higher than those of the differentiating cells in 2D monolayers (Fig. 1B). A Wnt/ꞵ-catenin pathway target gene, AXIN2, and a negative regulator of the Wnt/ꞵ-catenin pathway, DKK1, showed peak expression at D2 and decreased at D4 in both differentiation systems (Fig. 1B). Consistent with mesoderm and cardiac specification, ISL1, a cardiac progenitor marker, and MEF2A and MEF2C, transcription factors for cardiovascular development, were extensively expressed on D4 in differentiating cells in 3D culture (Fig. 1B). On D24 the cardiac differentiation and maturation was completed in both 2D and 3D culture systems, which successfully and stably retained a spontaneous beating property (Fig. 1C, Videos S1 and S2). In flow cytometry analysis on D24, the cTnT-positive population showed similar differentiation yields in both differentiation systems (2D-CMs; ±94.3%, 3D hiCOs; ±96.6%) although the purification step was not applied to the 3D differentiation system (Fig. 1C). However, the mRNA expression level of TNNT2, which encodes cTnT, was significantly higher in 3D hiCOs than 2D-CMs (Fig. 1D). These results together suggested that the spatiotemporal environment of 3D differentiation resulted in enhanced mesodermal and cardiac differentiation of hiPSCs, although the same modulation of the Wnt/ꞵ-catenin pathway was given in the early differentiation stage in both the 2D and 3D differentiation systems.
2. Three-dimensional cardiac organoids exhibit a mature sarcomeric isoform profile
Generally, the beginning of sarcomere assembly is the outstanding feature of the cardiac differentiation process, and new sarcomeres continuously elongate by alignment with thin filaments through massive expansion of myofibrils, leading to sarcomeric isoform switching [25]. Therefore, we analyzed the isoform switching in 2D-CMs and 3D hiCOs based on the gene expression level of cardiac myofibril assembly-related isoforms, which are a sign of sarcomere maturation. In the developing heart, the predominant isoform of myosin heavy chain (MHC) protein switches from β-MHC (MYH7) to α-MHC (MYH6) [25–28], and mature CMs express a higher level of cardiac troponin I3 (TNNI3) than fetal cardiac troponin I1 (TNNI1) [25,29–35].
Although the ratio of the ventricular type of cardiac cells compared to the atrial type (assessed by the expression ratio of MYL2 and MYL7) gradually increased during cardiac differentiation in both the 2D and 3D differentiation systems, the 3D hiCOs highly expressed MYH6 and TNNI3 rather than the fetal cardiac marker, MYH7 and TNNI1, in the intermediated (D8 and D16) stage as well as in the late stage (D24) of differentiation (Fig. 2A). Synthetically, this myofibril maturity through sarcomere isoform switching reinforces the idea of a higher maturation of 3D hiCOs during cardiac differentiation. The protein profiles regarding maturation of the sarcomere structure were confirmed by western blot analysis at D24 after cardiac differentiation. The 3D hiCOs showed higher expression of cTnT (encoded by TNNT2), the cTnI isoform (encoded by TNNI3), and ventricle-specific MYL2V (encoded by MYL2), but no significant expression of MYL2A (encoded by MYL7) compared with the 2D-CMs (Fig. 2B). These results also suggest robust cardiac structural maturation in the 3D hiCOs.
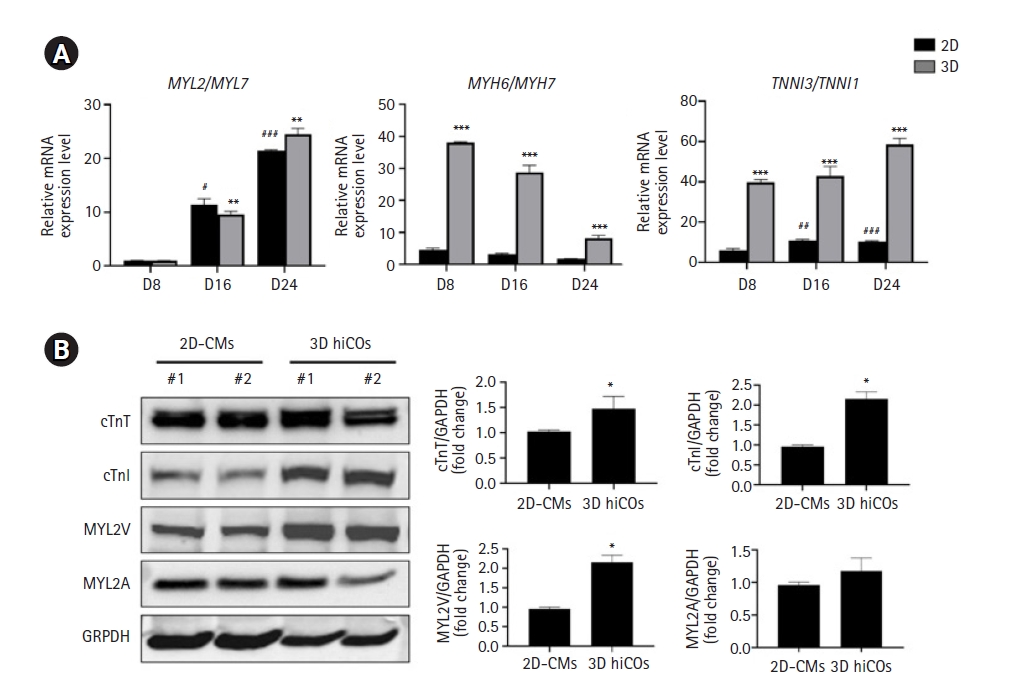
Human induced pluripotent stem cell-derived cardiac oragnoids (hiCOs) reveal a faster cardiac maturation phenotype. (A) The ratios of the relative expression of MYL2:MYL7, MYH6:MYH7, and TNNI3:TNNI1 genes in 2-dimensional (2D)-cardiomyocytes (CMs) and 3-dimensional (3D) hiCOs on D8 to D24. The expression of each gene was quantified by the comparative 2−ΔΔCT method and normalized to GAPDH, and then their relative ratio was calculated. (B) The protein expression levels of sarcomeric isoforms were analyzed using the western blot method. Equal protein loading amounts were confirmed by GAPDH expression. The corresponding density ratio was calculated by the average intensity of the bands from ImageJ software. The statistical analysis were conducted using the multiple t-test for 3 independent experiments (##p<0.01 and ###p<0.001 versus 2D human induced pluripotent stem cells [hiPSCs] at D0; n=3, **p<0.01 and ***p<0.001 versus 2D hiPSCs at each day; n=3). cTnT, cardiac troponin T; cTnI, cardiac troponin I.
3. Three-dimensional cardiac organoids express increased cardiac-specific ion channels and connexin-43
To determine differences in gene expression associated with the electrophysiological properties of 2D-CMs and 3D hiCOs, we evaluated the expression profile of several cardiac-specific ion channel genes during the intermediate (D8 and D16) to late (D24) stage of CM differentiation. The gene SCN5A, which encodes a protein involved in sodium influx and membrane depolarization in phase 0 of the cardiac action potential, remarkably increased from D8 to D24 in 3D hiCOs (Fig. 3A). The CACNA1D gene, which encodes the voltage-dependent L-type calcium channel (LTCC) in phase 1 of the action potential, also showed higher expression in 3D hiCOs than in 2D-CMs (Fig. 3A). However, the gene encoding a major calcium voltage channel of CM, CACNA1C, showed a similar gene expression increase in 2D-CMs and 3D hiCOs at D16 and D24 (Fig. 3A). Increased expression of RYR2 (calcium release channel in the sarcoplasmic reticulum) and KCNQ1 (Kv7.1 involved in repolarization phase [36]) was also observed in intermediate (D8 and D16) to late CMs (D24) in both the 2D and 3D culture systems; however, the 3D hiCOs showed more abundant gene expression than the 2D-CMs, with decreased expression of HCN4, a pacemaker-specific gene (Fig. 3A), associated with the reversal of maturational changes in human CMs [37,38]. These results suggest a higher maturity of the functional cardiac ion channel expression profile in 3D hiCOs than in 2D-CMs. Furthermore, we analyzed the protein expressions of connexin-43 (CX43) and Cav1.2, which are essential for cardiac excitation and contraction. The robust expression of CX43 was observed in 3D hiCOs (Fig. 3B), and the expression of Cav1.2 was also significantly higher in 3D hiCOs than in 2D-CMs (Fig. 3B).

The relative expression profile of cardiac-specific ion channels and connexin-43 (CX43). (A) Real-time quantitative polymerase chain reaction analysis of cardiomyocyte (CM) differentiation in 2-dimensional (2D)-CMs and human induced pluripotent stem cell-derived cardiac oragnoids (hiCOs). The relative expression levels of cardiac-specific ion channels regarding to action potential kinetics were studied at intermediate (D8 and D16) and late stages of cardiac differentiation (D24). The expression of each gene was calculated according the comparative 2−ΔΔCT method and normalized as the fold change based on the corresponding 2D-CMs on each day. (B) Western blot analysis in cell lysates from 2D-CMs and 3-dimensional (3D) hiCOs. The protein expression of CX43 and Cav1.2, which are essential for cardiac excitation and contraction were studied at late stage of cardiac differentiation (D24). Equal protein loading amounts were confirmed by GAPDH expression. The corresponding density ratio was calculated by the average intensity of the bands from ImageJ software. The statistical analysis was conducted using the multiple t-test for 3 independent experiments (#p<0.05, ##p<0.01 and ###p<0.001 versus 2D hiPSCs at D0; n=3,*p<0.5, **p<0.01, and ***p<0.001 versus 2D-CMs at each day; n=3).
4. Three-dimensional cardiac organoids present strong Ca2+ transient and contractility properties
With the results of enhanced cardiac specification and maturation of cardiac differentiation in 3D culture, we next determined the intracellular Ca2+ transients in 2D-CMs and 3D hiCOs at D24 of differentiation. After the loading of Calcium 6 green dye (a green fluorescent Ca2+ indicator) in each culture, spontaneous beating in both 2D-CMs and 3D hiCOs was observed using a Nikon laser-scanning fluorescence microscope (Fig. 4A), and the signal was simultaneously recorded by a microplate reader. A representative fluorescence image displayed the 3D hiCOs with synchronized strong Ca2+ transients with stable beating, whereas 2D-CMs showed relatively weak Ca2+ transients with scattered beating in a partial area (Fig. 4A, Videos S3 and S4). Each observed Ca2+ influx was quantified by intensity values in a microplate reader. The fluorescence intensity in 2D-CMs showed well-to-well variation during signal measurements, but that in 3D hiCOs was relatively stable (Fig. 4B). A calculation of the signal amplitude and beating rate of 2D-CMs and 3D hiCOs based on the fluorescent intensity revealed that the signal amplitude was twice as strong in the 3D hiCOs than in the 2D-CMs (Fig. 4C), and the 3D hiCOs exhibited an increased beat frequency that was close to the normal human heartbeat rate (about 60 beats per minute) (Fig. 4D) and had a rapid time-to-peak (Fig. S1).
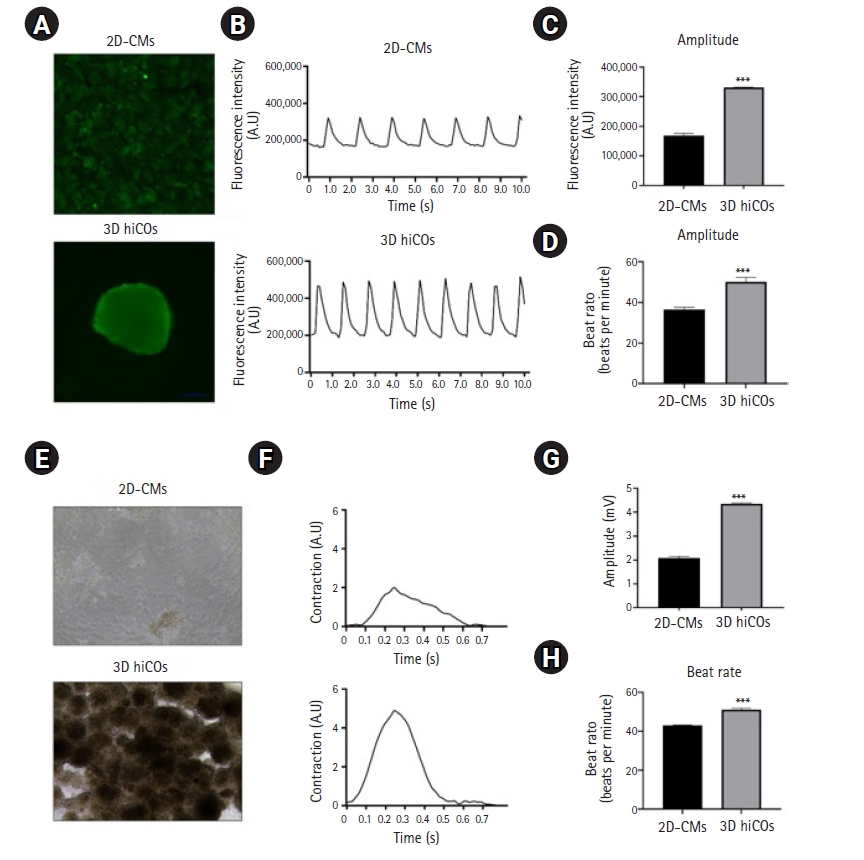
Human induced pluripotent stem cell-derived cardiac oragnoids (hiCOs) display functional maturation of Ca2+ transients and contraction properties. (A) Representative green fluorescence images were captured by a Nikon laser-scanning fluorescence microscope and the calcium transient was monitored on SpectraMAx iD3 multi-mode microplate readers (×200 magnification). (B) The real-time calcium influx was exported as the absolute value of fluorescence intensity (A.U.) in FLIPR Calcium 6 dye (green)-loaded 2-dimensional (2D)-cardiomyocytes (CMs) and 3D hiCOs. (C) The peak amplitudes of fluorescence transients were calculated by subtracting the minimum [Ca2+] from the maximum [Ca2+] in 2D-CMs and 3D hiCOs. (D) Each beat rate (beats per minutes, BMP) was converted from the represented value of fluorescence intensity (A.U.) in (A) by a series of mathematical calculations. (E) Representative optical images were captured from cells cultured on FLEXcyte 96 plates, and their typical signals were recorded on FLEXcyte 96 equipment once every 5 minutes for 60 minutes. Each graph for normalized amplitude (mV) was depicted by representative recording traces. (F) Each amplitude (mV) and (G) each beat rate (BPM) were converted from the represented trace values through an in-lab-developed calculation method. The statistical analysis was conducted using the multiple t-test for 3 independent experiments. (H) Beat rate (BPM) was converted from represented trace value through an in-lab-developed calculation method. The statical analysis were conducted by multiple t-test of three independent experiments (*p<0.5, **p<0.01, and ***p<0.001 versus 2D-CMs; n=3).
For a better understanding of in-depth cell functionality regarding the contraction force in 2D-CMs and 3D hiCOs, we performed a FLEXcyte 96 cardiac contractility assessment. Representative optical images displayed large clusters of multiple 3D hiCOs that exhibited strong contraction force and synchronized beating along with enhanced Ca2+ transients compared with 2D-CMs (Fig. 4E, Videos S5 and S6). Each observed signal was quantified by its value on a particular trace (Fig. 4F), and the 3D hiCOs presented higher amplitudes and beat rates than the 2D-CMs (Fig. 4G–H). Synthetically, these results showed that 3D culture leads to the maturation of Ca2+ transient properties in hiPSC-derived CMs with improved contractile function compared to 2D culture.
Discussion
hiPSC-CMs have provided the first and easily approachable equivalent of native human CMs. However, as knowledge of hiPSC-CMs has been gathered, several concerns have been raised because of the immature structure and function of hiPSC-CMs [39–41]. To overcome these limitations, researchers have attempted to mimic the in vivo heart environment using techniques such as a prolonged culture period, co-culture with other cells, and mechanical, electrical, and chemical engineering to increase the functionality of hiPSC-CMs [20,42–46]. Three-dimensional differentiation including organoids has been proposed as another maturation strategy, because it allows the generation of models resembling normal embryogenesis and cardiogenesis, with the realization of different signals and spatial gradients in the tissue microenvironment [47].
In this study, we directly compared the differentiated CMs from 2D and 3D culture systems and unveiled different cardiac differentiation patterns and functionality in each differentiation system. Although the yield of cardiac differentiation was not impacted by the culture system, we demonstrated that 3D hiCO differentiation facilitated early cardiac differentiation, switching myofibril assembly toward the mature phenotype, and increased cardiac ion channel expression, with stable and strong contraction of hiPSC-derived CMs.
Importantly, ion channels are critical for all aspects of cardiac maturation and functions by regulating cardiac action potential, calcium handling kinetics, and contractility [48–51]. In 3D hiCOs, expression of the SCN5A gene encoding voltage-gated sodium channels (mainly Nav1.5) more significantly increased during differentiation than in 2D-CMs, which explains why action potentials could be quickly initiated by rapid channel opening of the channel in the 3D environment. Consistent with higher cell excitability governed by Nav1.5, which is abundant in the cell-cell coupling of 3D hiCOs owing to a robust increase in CX43 membrane expression levels, consequently led to tightened cell coupling and facilitated impulse propagation [52]. From the cardiac structural point of view, the pronounced expression of CX43 and RyR2 could have important implications concerning intact myofibril assembly and sarcoplasmic reticulum function during structural maturation in 3D hiCOs [53].
hiPSC-derived CMs are known to be devoid of the sophisticated adult CM structure, T-tubules [54]. Structural alignment of the calcium channel on T-tubules and RyR2 on the sarcoplasmic reticulum is crucial for dyad formation, which plays an important role in efficient excitation-contraction coupling and Ca2+-induced Ca2+ release [55]. Despite the lack of obvious observations of co-localization between calcium channels and RyR2, the significantly higher gene and protein expression of these 2 channels in 3D hiCOs are in excellent agreement with their enhanced functionality, as confirmed by calcium transient and contractility measurements. The clustering of RyRs may link with the abundant calcium channels in 3D hiCOs, resulting in the efficient increase in [Ca2+]i and strong contraction force with a more rapid time-to-peak and enhanced signal amplitude compared with that in 2D-CMs. Moreover, cardiac excitation and contraction depend on specific ventricular ion channels, including LTCC (such as Cav1.2) and CX43 localized in the CM plasma membrane, which are essential for beat-to-beat heart function to generate contractile force [56]. Our results have also verified the gene and/or protein expression profiles in parallel with these 2 ion channels (Fig. 3). These findings may suggest that 3D hiCOs are likely to be structurally and functionally more mature than 2D-CMs.
In conclusion, we differentiated hiPSCs into CMs using temporal modulation of the Wnt/ꞵ-catenin pathway signaling and compared structural and functional differences between 2D monolayer and 3D organoid culture. Through a comparison of these differentiation systems, we ultimately revealed a high enrichment of mature CMs in 3D cardiac organoid formation. Thus, this study could aid in the basic understanding of cardiac maturation research in culture according to the topological environment of cardiac differentiation and provide relevant evidence regarding the potential use of hiPSC-derived cardiac organoids as an alternative way to verify organ behavior and disease mechanisms, or to increase the efficiency of drug screening and drug safety/toxicology.
Notes
Conflict of interest
No potential conflict of interest relevant to this article was reported.
Funding
This study was supported by grants from the National Research Foundation funded by the Ministry of Science and ICT of Korea (No. 2019M3A9H1103718 and 2022M3A9H1014160).
Author contributions
Conceptualization: HL, DHW; Data curation: HL, DBC, JSI; Methodology: HL, JSI; Formal analysis and visualization: HL, HW; Validation: JA, SBK, SYe, SYo; Writing-original draft preparation: HL, Writing-review and editing: JA, DHW.
Supplementary Material
Supplementary materials are presented online (available at https://doi.org/10.51335/organoid.2022.2.e14).
Videos S1.
Representative optical beating video of hiPSC-derived cardiomyocytes at D24, related to Figure 1(C). This video was recorded at 100x magnification (https://doi.org/10.51335/organoid.2022.2.e14.v01).
Videos S2.
Representative optical beating video of hiPSC-derived cardiac organoids cultured for 24 days in ultra-low attachment plates, related to Fig. 1C. This video was recorded at 40x magnification (https://doi.org/10.51335/organoid.2022.2.e14.v02).
Videos S3.
Calcium 2+ ion transients in a showing electrical coupling of 2D-cardiomyocytes, related to Figure 4 (A). The scale bar represents 100 μm (https://doi.org/10.51335/organoid.2022.2.e14.v03).
Videos S4.
Calcium 2+ ion transients showing electrical coupling by beating of hiCOs, related to Figure 4 (A). The scale bar represents 100 μm (https://doi.org/10.51335/organoid.2022.2.e14.v04).
Videos S5.
2D cardiomyocytes beating at day 7 on FLExcyte 96 plate, related to Figure 4 (E). This video was recorded at 100x magnification (https://doi.org/10.51335/organoid.2022.2.e14.v05).
Videos S6.
3D cardiac organoid beating at day 7 on FLEcyte 96 plate, related to Figure 4 (E). This video was recorded at 40x magnification (https://doi.org/10.51335/organoid.2022.2.e14.v06_.
Fig. S1.
Measurement of Time-to-peak by Calcium transient analysis. Representative fluorescence time-lapse calcium imaging in 2-dimensional (2D)-cardiomyocytes (CMs) and 3D hiCOs loaded with the intracellular calcium indicator FLIPR Calcium 6 dye (green) weas monitored during 10 seconds. The real-time quantitative intensity value was exported in Excel software and time-to-peak were converted through an in-lab-developed calculation method from 3 independent experiments. The statical analysis were conducted by multiple t-test of 3 independent experiments (***p<0.001 versus 2D-CMs; n=3).

