A method for culturing patient-derived lung cancer organoids from surgically resected tissues and biopsy samples
Article information
Abstract
Background
The cancer model system maintaining genetic and phenotypic characteristics of human cancers is crucial for the study of precision cancer medicine. In this respect, patient-derived cancer organoids have been developed as a preclinical cancer model with significant advantages over previous cancer models including PDX and cell lines. Patient-derived cancer organoids are generated through the process of self-organization of cancer cells in the tissue tissues, resulting in recapitulating phenotypic and genetic attributes of their parental tissues. For this reason, cancer organoids can be used in drug development and personalized medicine. Therefore, the development of effective cancer organoid culture methods is required in the cancer research field.
Methods
All lung cancer samples were obtained from Asan medical center (Seoul, South Korea) with consent from patients. For this study, lung cancer tissues were obtained by surgery and EBUS-TBNA. To generate LCOs from these samples, we used two different methods according to the source of tissues.
Results
This method improves the efficiency of LCOs setups composed of pure cancer cells and describes an additional procedure for reconstructing LCOs after cryopreservation. We confirmed that stock LCOs had the ability to self-organization and retain the morphological and genetic characteristics of their parental tissues. They also maintain their responsive property to certain anticancer drugs after thawing.
Conclusion
This study described a detailed protocol for establishing LCO from surgically resected tumor tissues and endoscopic biopsy samples. Also, we demonstrated that the LCO model generated by using these methods enables anticancer drug screening at the individual patient level.
Introduction
Cancer model systems are essential for cancer research. For precision cancer medicine, model systems must reflect the characteristics of genetic and phenotypic heterogeneity in human cancers. Patient-derived cancer organoids can be generated from human cancer tissues through the process of self-organization of cancer cells in the tissue [1]. Through this process, cancer organoids recapitulate the phenotypic and genetic attributes of the original patient tumor tissues. These characteristics are preserved during long-term culture [2]. Therefore, patient-derived cancer organoids are expected to be used to predict the clinical response of individual patients to anticancer therapy.
Lung cancer is extremely heterogeneous, and personalized cancer models are increasingly important to overcome the most lethal cancers. Traditionally, lung cancer is classified into 2 main histological classes—small cell lung cancer and non-small cell lung cancer—based on tumor cell morphology. Non-small cell lung cancer is categorized into several subtypes, including adenocarcinoma, squamous cell carcinoma, large cell carcinoma (neuroendocrine and non-neuroendocrine), and adenosquamous carcinoma [3]. The standard treatment for early-stage lung cancer is surgical resection, whereas advanced-stage lung cancer is treated with tumor type-tailored therapy or genotype-based treatment according to the pathologic classification and molecular testing, as well as surgery [2]. Since tumor characteristics, including the histological subtype, are major factors to consider for the treatment of lung cancers [4], a cancer model system covering various histologic subtypes is required. In this respect, patient-derived cancer organoids are the most representative models of lung cancer, which is highly heterogeneous. In clinical practice for lung cancer, tissue samples for generating organoids can be obtained by surgery or various biopsy procedures, including endobronchial ultrasound (EBUS)‑guided transbronchial needle aspiration (TBNA) [5].
In this paper, we describe a method for the generation of lung cancer organoids (LCOs) by using patient samples obtained from surgery and EBUS-TBNA. We also share LCO culture know-how by describing our failures. The LCOs that we generated stably recapitulated individual patient cancer characteristics, including morphological and genetic features, even after cryopreservation. The LCOs were tested to predict their response to targeted drugs based on genomic features. Given the results, the LCO model generated using our methods is a potential personalized cancer model to predict the response of an individual patient to anticancer therapy.
Materials and Methods
Ethics statement: This study was approved by the Institutional Review Board of Asan Medical Center (No. #2018-0152).
1. Human specimens and organoid culture.
All samples used in this study were provided by the Asan Bio-Resource Center, Korea Biobank Network (2018-25(179)). The research protocol was approved by the Institutional Review Board of Asan Medical Center (#2018-0152; Seoul, Republic of Korea). The entire experimental protocol was conducted in compliance with the institutional guidelines.
Surgically resected samples (approximately 1–4 cm3) and biopsy specimens (~1×30 mm) obtained from EBUS-TBNA were transported to the laboratory on ice within 1 hour of removal from patients in cold Hank’s balanced salt solution (HBSS) with antibiotics (Lonza, Basel, Switzerland). Samples were washed 3 times with cold HBSS that contained antibiotics. The surgically resected tissue was incubated with 0.001% DNase (Sigma-Aldrich, St. Louis, MO, USA), 1 mg/mL collagenase/dispase (Roche, Basel, Switzerland), 200 U/mL penicillin, 200 mg/mL streptomycin, and 0.5 mg/mL amphotericin B (2% antibiotics; Sigma-Aldrich) in DMEM/F12 medium (Lonza) at 37°C for 1 hour with intermittent agitation. The biopsy sample was incubated with TrypLE Express Enzyme (Gibco, Waltham, MA, USA) at 37°C for 5 minutes with intermittent agitation and treated with red blood cell (RBC) lysis buffer (Sigma-Aldrich) for 5 to 10 minutes at room temperature. After enzymatic digestion, the suspensions were repeatedly triturated by pipetting. The cells were centrifuged at 112×g for 3 minutes, and the pellet was resuspended in 50 μL of mininal basal medium (MBM) (serum-free medium (DMEM/F12; Lonza) supplemented with 20 ng/mL basic fibroblast growth factor (Invitrogen, Waltham, MA, USA), 50 ng/mL human epidermal growth factor (Invitrogen), N2 (Invitrogen), B27 (Invitrogen), 10 μM ROCK inhibitor (Enzo Life Sciences, New York, NY, USA), and 1% penicillin/streptomycin (Gibco) and 100 μL of Matrigel. The resulting cell suspension was allowed to solidify on pre-warmed 6-well culture plates (Corning, Corning, NY, USA) at 37°C for 10 minutes. After gelation, 3 mL of MBM was added to the well. The medium was changed every 4 days.
For passaging, a solidified Matrigel drop containing the organoids was harvested using cold Dulbecco’s phosphate-buffered saline (DPBS) and centrifuged at 1,000 rpm for 3 minutes at 4°C. The pellets were washed with cold DPBS and centrifuged at 1,500 rpm for 15 minutes at 4°C. The organoids were resuspended in 2 mL of TrypLE Express (Gibco) and incubated for 10 minutes at 37°C for dissociation. Later, 10 mL of DMEM/F12 containing 10% fetal bovine serum (FBS) was added, and the samples were centrifuged at 1,000 rpm for 3 minutes. The pellets were washed with DPBS and centrifuged at 1,000 rpm for 3 minutes. The pellets were resuspended in MBM+Matrigel (1:3) and reseeded at 1:3 to 1:4 ratios to allow the formation of new organoids.
To obtain stored LCOs, LCOs cultured in 3 to 4 wells of a 24-well plate were harvested using cold DPBS and then centrifuged at 1,000 rpm for 3 minutes at 4°C. The pellets were washed with cold DPBS and centrifuged at 1,500 rpm for 15 minutes at 4°C. The supernatant was removed and the LCO pellet was resuspended in freezing medium: Culture Media 7: ES grade FBS (Gibco) 2: DMSO (Sigma-Aldrich) 1. The vials for storage were transferred to a deep freezer, followed by a gradual reduction in temperature to −80°C, and transferred to vapor-phase liquid nitrogen storage.
2. Immunohistochemistry
Tissues and organoids harvested in 4 to 6 wells of a 24-well plate (>passage 3) were fixed in 4% paraformaldehyde followed by dehydration, paraffin embedding, sectioning, and standard hematoxylin and eosin (H&E) staining. Nuclei were counterstained with Harris hematoxylin. Images were acquired on a Leica Eclipse E600 microscope.
3. Genomic analysis
DNA was extracted from tumor tissues with matched normal tissues and early-, intermediate-, and late-passage LCOs from AMC 15LT-005 and 006 lines using the DNeasy Blood & Tissue Kit (Qiagen, Hilden, Germany) according to the manufacturer’s protocol. In order to evaluate the somatic mutations in the paired tumor tissues and early-, intermediate-, and late-passage LCOs, Whole-exome sequencing was performed using the Illumina Hiseq 2500 platform. A SureSelect Exome Enrichment kit version 6.0 (Agilent Technologies, Santa Clara, CA, USA) was used to capture all exome regions for whole-exome sequencing. The DNA libraries were sequenced for 150-bp paired-end sequencing. The Illumina sequencing data were processed using the Genome Analysis Toolkit (GATK) v1.6.5 [6]. The sequenced reads were aligned to the human reference genome (NCBI build 37) with the Burrows-Wheeler Aligner (ver. 0.5.9) [7] using the default options. De-multiplexing was performed using MarkDuplicates of the Picard package to remove polymerase chain reaction duplicates. De-duplicated reads were realigned at known indel positions with the GATK IndelRealigner, and the base quality was recalibrated using the GATK TableRecalibration [6,8]. Somatic variant calling for single-nucleotide variants and short indels was performed with matched normal tissues using MuTect (v1.1.7) [9] and SomaticIndelocator in GATK, respectively. Germline variants from somatic variant candidates were filtered out with common dbsnp (build 141; found in ≥1% of samples), the Korean Reference Genome Database, and a panel of normal samples. The final somatic variants were annotated using the Variant Effect Predictor (v79) and then converted to a mutation annotation file format using vcf2maf. Quality checks for fastq files were performed using FastQC (http://www.bioinformatics.babraham.ac.uk/projects/fastqc/). Quality checks for analysis-ready BAM files, including the total read number, percentage of pass-filter reads, percentage of selected bases, mean target depth, percentage of target not covered, percentage of target bases covered 30X, and the percentage duplication for cleaned BAM files was performed using the CalculateHSMetrics and MarkDuplicates modules of the Picard program. Integrative Genomics Viewer was used to view the BAM files [10]. R software (v3.0) with the maftools [11], ComplexHeatmap, and ggplot2 packages were used for data visualization. Fingerprinting analysis was performed using the panel detecting 24 SNP alleles on the Sequenom MassARRAY technology platform (Sequenom, CA, USA).
4. Drug screening
LCOs cultured in 6 to 12 wells in a 24-well plate over 2 weeks were harvested and dissociated using TrypLE Express (Gibco). The dissociated LCOs were counted and seeded to 10 μL of 2×103 cells per well on a 96-well white plate (Corning) after being mixed in MBM+Matrigel (1:3 ratio). After gelation, 100 μL of MBM was added to each well. The LCOs were allowed to grow to a diameter of 60 to 70 μm. Next, gefitinib (Selleck Chemicals, Houston, TX, USA) was administered in 10 concentration ranges every 3 days in triplicate. DMSO treatment was applied as a control. After 6 days, the medium was changed to 100 μL of MBM per well to measure cell viability, and 100 μL of CellTiter-Glo (Promega, Madison, WI, USA) was added to each well. The plates were agitated for 30 minutes at room temperature prior to luminescence reading. IC50 values were determined using GraphPad Prism (v5.01; GraphPad LLC, San Diego, CA).
Results
1. The isolation of lung cancer cells from surgically resected tissues and biopsies
To culture LCOs, we used small fractions (approximately 1–4 cm3) of resected tissues through video-assisted thoracoscopic surgery lobectomy and biopsy samples obtained from EBUS-TBNA. As the 2 types of samples were different in terms of amount and condition, we used different initial steps for each sample type (Fig. 1A).
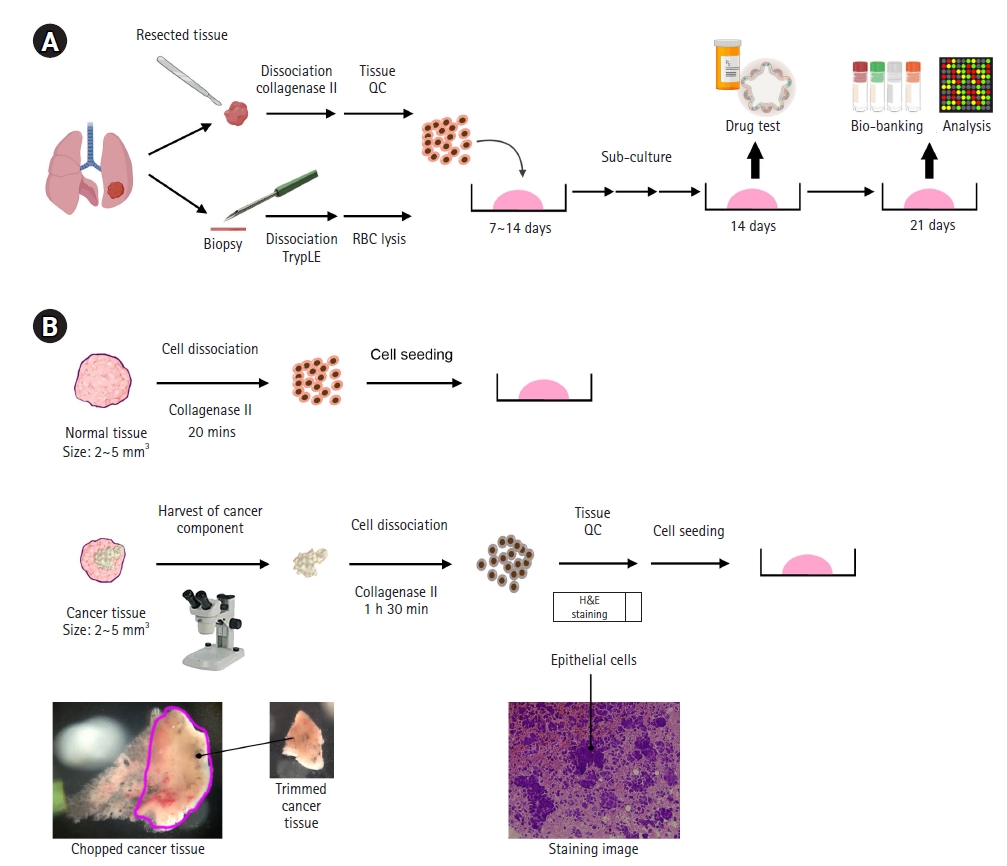
The generation of lung cancer organoids (LCOs) from surgically resected tissues and biopsy samples. (A) A schematic illustration of the procedure from LCO establishment to biobanking and analysis. (B) The initial steps for improving the LCO generation rate, when using surgically resected tissues. QC, quality check; RBC, red blood cell.
For fresh cancer tissues obtained after surgical resection, we chopped the tissue into tiny pieces (approximately 2–5 mm2) and trimmed the tissue under microscopy to obtain the area with the highest cancer purity (Fig. 1B). Collagenase type II was used to dissociate these tissues to obtain cells. Before seeding these cells, we performed H&E staining to test the cell composition in these tissues (Fig. 1B). Only samples in which epithelial cells were observed were cultured to cancer organoids. In addition, we obtained fresh non-malignant tissue together with the cancer tissue at the same time. We chopped this non-malignant tissue into tiny pieces (approximately 2–5 mm2) and dissociated the tissue into cells by treating it with collagenase type II for 20 minutes (Fig. 1B). We then seeded 5×105–2×106 cells with Matrigel (Corning) into each well of a 6-well plate and cultured these cells for 7 to 14 days (Fig. 2A, passage 0; P0).
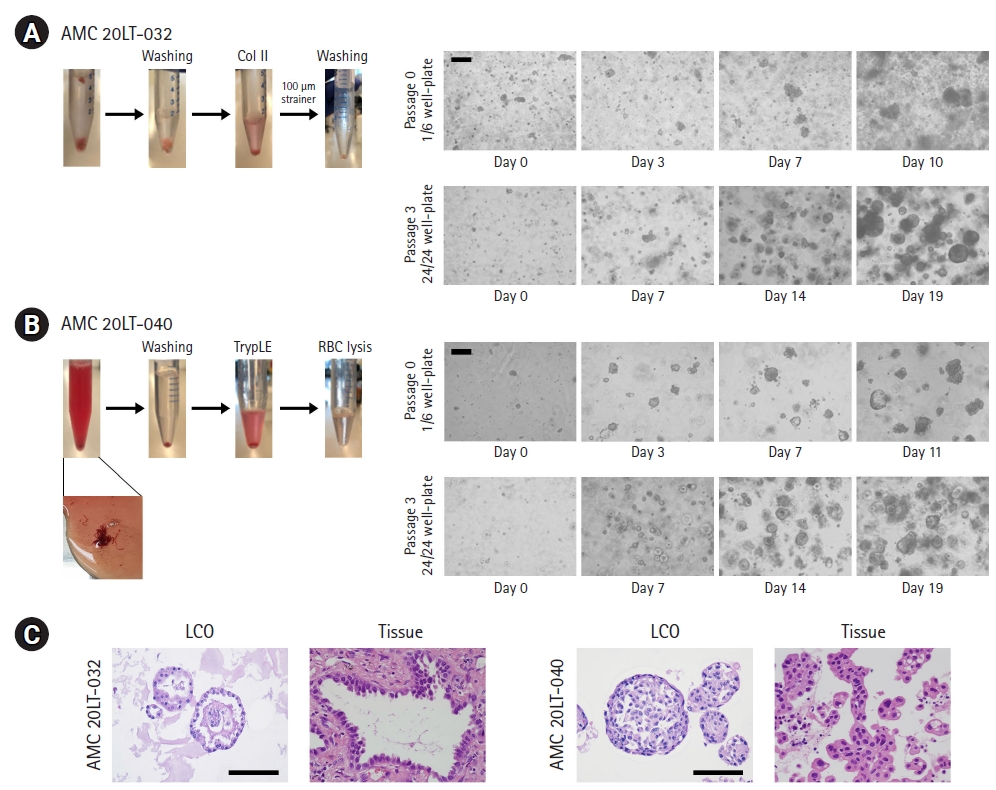
The cultured lung cancer organoids (LCOs) represented the morphological features of original patient tissues. (A, B) Representative images of patient samples and LCOs cultured at passages 0 and 3. The LCOs in these images were as follows: AMC 20LT-032 obtained from surgically resected tissue (A), invasive adenocarcinoma; AMC 20LT-040 obtained from EBUS-TBNA biopsy (B), metastatic adenocarcinoma. Scale bar, 200 μm. (C) H&E staining images of LCOs and their original cancer tissues. Scale bar, 100 μm.
For lung cancer biopsies obtained from EBUS-TBNA, we directly added TrypLE into the tube containing the biopsy sample. As shown in Fig. 2B, the biopsy samples contained more blood than the surgically resected samples. For this reason, after treatment with TrypLE for 5 minutes at 37°C, we removed the RBCs from the biopsy using RBC lysis buffer for 5 to 10 minutes at room temperature (Fig. 1A and Fig. 2B). We then seeded the cells with Matrigel (Corning) in one well of a 6-well plate and cultured these cells for 7 to 14 days (Fig. 2B, passage 0; P0).
2. The generation of LCOs
During P0, cell clusters formed round shapes within 5 days. Organoid morphology was observed after 7 days (Fig. 2A and 2B). Generally, the samples obtained from EBUS-TBNA contained a lower confluence of cancer cells than surgically resected samples. Therefore, after seeding, the cancer cell numbers in these samples were lower than those in surgically resected samples (Fig. 2A and 2B). When the confluence of organoids reached about 80% and the diameter of organoids was an average of 150 to 200 μm, we performed sub-passaging using these organoids. For passaging, organoids were dissociated using TrypLE for 20 minutes at 37°C. The organoids were split into small cell clusters or broken down into single cells. We seeded these cells at 2×104 cells per well of a 24-well plate and cultured these cells for 14 to 21 days (Fig. 2A and 2B). After passage 3, we validated the similarity between the cultured organoids and their original tissues using H&E staining (Fig. 2C).
3. Case review of failed LCO culture
Most failed LCO cases showed fibroblast overgrowth. In case #1, the tissues were dissociated into cell clusters without problems related to the cancer cell population, and these cells were seeded in a Matrigel drop (Fig. 3A). After 3 days, they grew into organoids with round shapes. However, fibroblasts were observed on day 9, and organoids were deprived of their niches, as the fibroblasts grew faster than the organoids. Even though some organoids of middle size (average diameter, 100 μm) were observed, they mostly lost their niches at the next passage. Eventually, the growth of organoids was stopped (Fig. 3A).

Cases of failed lung cancer organoid (LCO) generation. (A) Representative images of LCOs showing the overgrowth of fibroblasts. (B) Representative images of LCOs cultured from individual cells. (C) Representative images of LCOs showing low self-organization. Scale bar, 200 μm.
Next, we observed that dissociating tissues into single cells hindered the generation of organoids. When most cells were dissociated into individuals instead of cell clusters, the cells took a long time to generate organoids or did not form pre-organoid structures (Fig. 3B).
We observed that some cases seeded as cell clusters did not completely generate organoids. As shown in Fig. 3C, only a few cells formed organoids (<10 organoids) in this case. This result indicated a low capacity for self-organization. Eventually, this case could not form organoids in the next passage (passage 1).
4. Anticancer drug testing according to the genetic features of LCOs.
To investigate the ability of stored LCOs to predict the outcomes of targeted therapy, 2 LCO stocks with EGFR mutations were selected, thawed, and cultivated [2]. As shown in Fig. 4A, the structure of the frozen LCOs was destroyed, but they soon reconstructed their structure during 3-dimensional in vitro culture. To investigate whether LCOs maintained their original cancer characteristics after being stored, we performed genomic analyses using DNA extracted from original tissue and LCOs before and after thawing. We stored LCO lines at early passage (passage <5; early-passage LCOs). After several months, we re-cultured LCOs after thawing (intermediate-/late-passage LCOs) and extracted DNA for investigating genomic changes. For the LCOs from AMC 15LT-005, we obtained DNA 3 times (passages 4, 10, and 16). The LCOs from AMC 15LT-006 were harvested 2 times (passages 4 and 13). Most of the mutations detected in original cancer tissues were maintained in early-, intermediate-, and late-passage LCOs. We demonstrated that our LCOs maintained the genetic characteristics of their original tissues after being reconstituted (Fig. 4B).
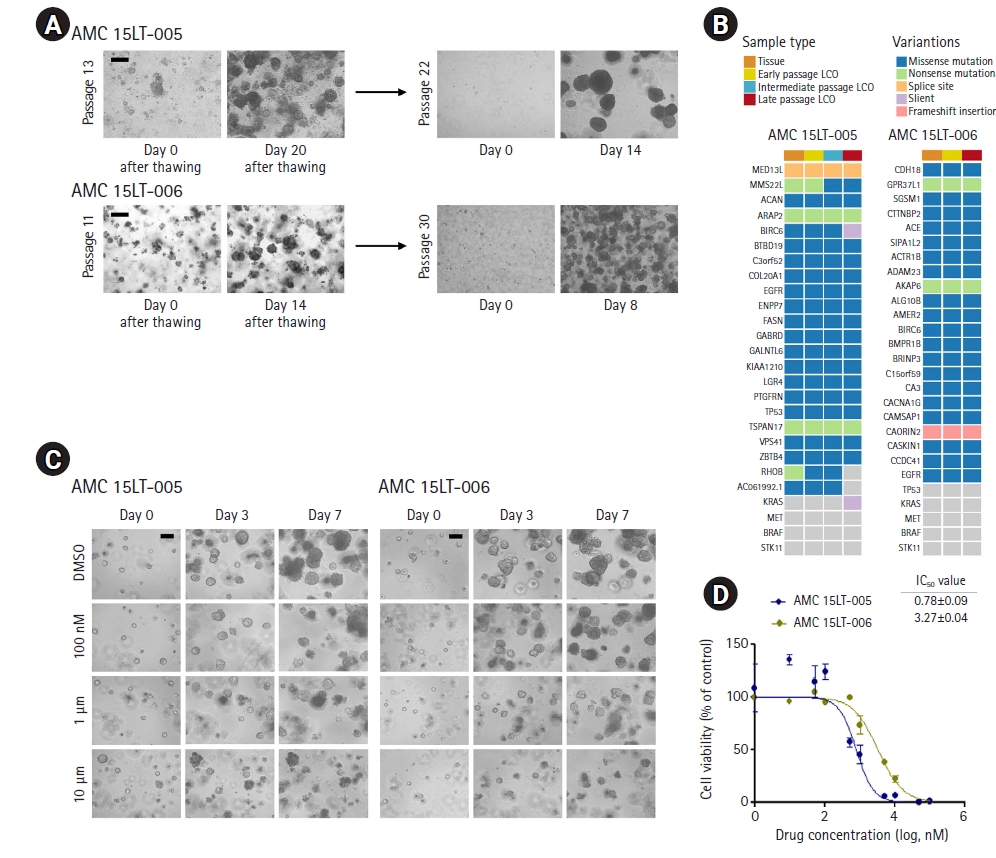
Stored lung cancer organoids (LCOs) have the potential to predict drug responses. (A) Bright-field microscopy images of stored LCOs after thawing. (B) Heatmap analysis of the 25 mutations in LCOs and their corresponding cancer tissues. (C) Bright-field microscopy images of LCOs showing the sizes and morphological changes of LCOs after treatment with DMSO or gefitinib at the indicated concentrations. (D) Dose-response curves after 6 days of treatment with gefitinib. Representative viability curves were generated from the luminescence signal intensities. Error bars indicate standard error of the mean (n=3). Scale bar, 200 μm.
Next, we treated the LCOS with gefitinib targeting the EGFRL858R mutation in these 2 LCO lines that maintained the mutation (Fig. 4B). For gefitinib treatment, we seeded 2×103 cells per well of a 96-well plate. When the diameter of organoids was an average of 60 to 70 μm, we administered gefitinib. As a result, growth inhibition and structural destruction in AMC 15LT-005 with EGFRL858R (reported as 15LT-43 in our previous paper [2]) were observed from day 3 after drug treatment (Fig. 4C). Meanwhile, AMC 15LT-006 with EGFRL858R/MET amplification (reported as 15LT-51 in our previous paper [2]) showed growth inhibition and structural destruction at a high concentration of gefitinib (Fig. 4C). To measure the IC50 value of each LCO after treatment with gefitinib, LCOs on day 6 after drug treatment were lysed the CellTiter-Glo lysis buffer (Promega). Next, we measured a luminescent signal proportional to the amount of adenosine triphosphate present in these LCOs. As expected, the IC50 value of AMC 15LT-005 (0.781 μM) was lower than that of AMC 15LT-006 (3.269 μM) (Fig. 4D). This result corresponds with our previous data [1]. Therefore, our stored LCOs are still available to predict patients’ drug responses.
Discussion
Cancer organoids are emerging as a new generation of in vitro preclinical cancer models, and have the potential to be used to predict individual patients’ clinical responses to cancer therapy [12,13]. However, cancer organoid technology shows several limitations, including the lack of standardized methods for some solid cancer organoids, challenges in producing cancer organoids within a clinically meaningful time window, and a relatively low success rate. By overcoming these limitations, organoid technology will be applied as a co-clinical track for clinical decision-making [13]. In this paper, we described an improved method for generating cancer organoids using several sample types. In addition, we showed that the LCO lines we generated can be used stably for anticancer drug tests after cryopreservation.
Lung cancer samples for the generation of LCO lines are obtained after surgery or endoscopy. In particular, specimens obtained from endoscopy including EBUS-TBNA are often collected prior to anticancer therapy [14]. Therefore, LCOs established from biopsies can be used to predict clinical drug responses. We have described a method to generate LCO lines from biopsies containing a high population of blood cells. In addition, we have described additional steps to improve the organoid success rate when using surgically resected tissues with various conditions. Additional steps, including trimming and tissue quality control, help to save time and expense in establishing organoid lines. Nevertheless, the success rate of LCO generation depends on the original cancer tissue characteristics. We have presented several failed cases in this paper to share various cases of lung cancer tissue. Fibroblast overgrowth is an obstacle to LCO culture. However, the role of fibroblasts in the cancer microenvironment cannot be ruled out, as it is associated with therapeutic effects [15]. Therefore, in order to study communication between cancer cells and fibroblasts, more research is needed to develop a culture method that regulates the growth of fibroblasts. When the tissues were dissociated into individual cells after collagenase treatment, the rate of LCO generation was low. In these samples, the ability to self-organize was found to be low. Therefore, obtaining small cell clusters rather than single cells helps to improve the generation rate of LCOs. Although a single type of medium was used for culturing LCOs, a customized LCO medium according to cancer tissue status would help to increase the success rate.
Targeted therapy according to genomic features has been widely used in the clinical treatment of lung cancer. Although targeted therapy has shown improved therapeutic effects, lung cancer remains an unmet need, with a recurrence rate of 30% to 50% after treatment [16]. Cancer organoids retain the molecular and functional characteristics of their parental tumors at the time of sample collection. Therefore, the storage and subsequent sharing of cancer organoids can provide important information such as alterations in drug responses and the identification of specific molecular signatures between primary and recurrent cancers [17]. In this respect, collections and biobanks of living cancer organoids are a valuable human bioresource. We demonstrated that stockpiled organoids retain donor genetic characteristics and can be used for drug screening. In general, methods for organoid cryopreservation require organoid samples with a freezing medium for liquid nitrogen storage, but the procedures have not yet been defined [18,19]. Cryopreserved organoids had compromised structures (Fig. 4A) and about 70% of cryopreserved LCOs were reconstituted with their original characters [2]. Therefore, a standardized method for the preservation of cancer organoids is needed for the stable storage of living human cancer resources.
In conclusion, we shared a method for generating LCOs from surgically resected tissues and biopsy samples, and demonstrated that our stored LCOs have the potential to be used in drug screening to predict drug responses in individual patients.
Notes
Conflict of interest
No potential conflict of interest relevant to this article was reported.
Funding
This study was supported by the National Research Foundation of Korea (NRF) grant funded by the Ministry of Science and ICT (MSIT, Korea) (NRF-2020R1C1C1004935 & 2019M3E5D3071926), and by the Technology Innovation Program for Fostering New Post-Genome Industry funded by the Ministry of Trade, Industry & Energy (MOTIE, Korea) (#10067796).
Author contributions
Conceptualization: MK, SJJ; Data curation: YK, MK; Formal analysis: YRK, JMK, YK, MK, SJJ; Funding acquisition: MK, SJJ; Investigation: YRK, JMK, MK, SJJ; Methodology: YRK, JMK, MK; Project administration: MK, SJJ; Resources: SJJ; Software: YK, MK; Supervision: MK, SJJ; Validation: MK, SJJ; Visualization: MK; Writing-original draft: MK; Writing-review & editing: MK, SJJ.
Data availability
Please contact the corresponding author for data availability.
