Toward brain organoid-based precision medicine in neurodegenerative diseases
Article information
Abstract
In recent years, the concept of precision medicine—an approach to developing personalized drugs for disease prevention or treatment—has emerged as an up-and-coming field in medical research. However, there are numerous limitations to applying this approach to the treatment of neurodegenerative diseases, such as Alzheimer disease, because of the invasiveness of obtaining human brain samples. Meanwhile, the development of human brain organoids has become one of the most powerful in vitro research tools because these organoids can be established from induced pluripotent stem cells or embryonic stem cells. In this review, we discuss how we can effectively utilize brain organoids for precision medicine research in conjunction with a variety of high-end techniques such as clustered regularly interspaced short palindromic repeats (CRISPR)-associated protein 9 (CRISPR-Cas9) genomic editing, integrative multi-omics analysis, 3D brain tissue clearing, and high-content screening confocal microscopy imaging systems. We herein provide new insights in order to materialize organoid-based precision medicine therapy and future directions.
Introduction
An organoid is a 3-dimensional (3D), self-organized tissue derived from human induced pluripotent stem cells (iPSCs) or embryonic stem cells [1]. In recent years, various types of stem cell-derived organoids including brain, gut, lung, and liver organoids, have been used for translational research because of their capacity to recapitulate biological functions in human organs, unlike other cell culture models [2–5]. Furthermore, given the fact that precision medicine therapy, an approach to develop personalized drugs for disease prevention or treatment, is an up-and-coming field in medical research, the optimal use of organoids is considered a promising aspect of precision medicine due to their expeditious genetic manipulation and their higher similarity to real human tissues than two-dimensional culture methods [6]. The applicability of cerebral organoids in neurodegenerative studies is especially essential for drug development due to the limited accessibility of the human brain. Brain organoids can be an alternative way to obtain personalized biological samples instead of directly opening the brain with a surgical biopsy. However, due to the difficulty in quality control of organoids (e.g., size variations of organoids, different cellular component proportions, and drug resistance to penetration, etc.) and the lack of strategies for narrowing-down candidate drugs, it is highly necessary to find proper methods for the development of organoid-based precision medicine platforms. Of course, many recent studies have tried to generate uniform organoids using microwell-containing plates or performed automatic quality controls for drug screening with high-content screening (HCS) systems [6–8]. Current bioinformatic methods have also opened a new era in computational data analyses, with mathematical network models for diseases, multi-omics analysis using different datasets, and the convergence of advanced validation processes with HCS systems for biological samples [6,9-11]. Nevertheless, both organoid-based drug-screening methods and systems-biological approaches still need to be developed and enrich each other.
Thus, we herein discuss how to materialize the development of organoid-based precision medicine for neurodegenerative diseases. We review current advanced technologies to modulate and improve organoid culture systems, such as clustered regularly interspaced short palindromic repeats (CRISPR)-associated protein 9 (CRISPR-Cas9) genomic editing [12], the generation of brain assembloids [13,14], and organ-on-a-chip methods. We also present perspectives for ranking candidate drugs against neurodegenerative diseases based on a combination of systems-biological approaches and organoid-based multi-omics data. Moreover, we describe current limitations barriers to the materialization of organoid-based precision medicine therapy and future directions.
Ethics statement: This study was a literature review of previously published studies and was therefore exempt from institutional review board approval.
Concept of organoid-based precision medicine
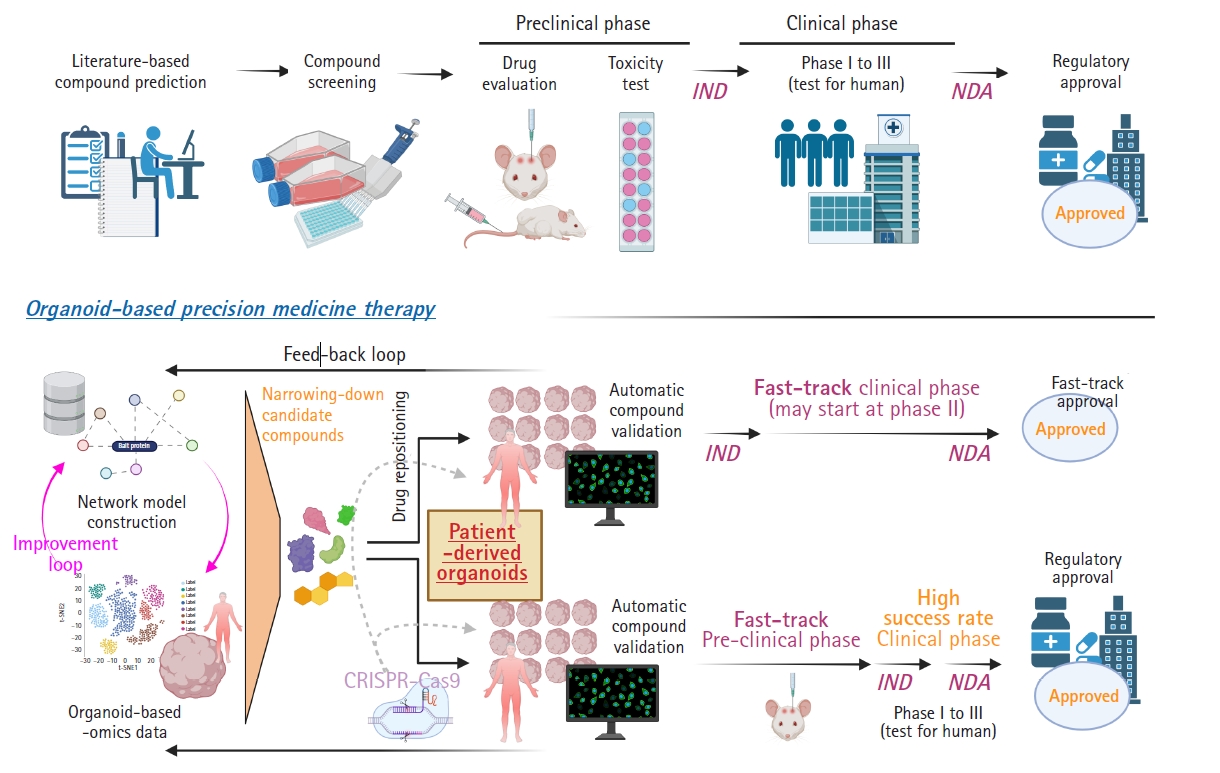
Organoid-based precision (or personalized) medicine is a new concept that utilizes human-derived organoid samples for drug-screening and, by extension, finding optimal candidate drugs through the convergence of systems-biological approaches (Fig. 1). First, in comparison with conventional drug discovery steps, organoid-based precision medicine can begin with different strategies for narrowing-down candidate drug compounds. Recent advances in systems biology-based mathematical models have made it possible to identify key disease mechanisms, which are validated with organoid-based omics data (e.g., the transcriptome, genome, proteome, and metabolome) instead of literature-based compound prediction [6]. Network models and organoid-based multi-omics data need continuous complements and feedback loops between them to improve the logicality of the model and incarnate a real human brain network. Next, a network of protein-ligand interactions and/or in silico perturbation analyses with disease-phenotype scoring can narrow the options down and make the drug testing steps more efficient [15,16]. During the validation steps, automatic compound validations using well-quality-controlled patient-derived organoids will be performed [6,17]. This step can include state-of-the-art tissue clearing methods and HCS systems. Moreover, CRISPR-Cas9-based genomic editing or drug repositioning strategies can be adopted to expedite the evaluation of medicines’ effects. Finally, the results of drug screening using patient-derived organoids can accelerate Food and Drug Administration (FDA) approval. Since current animal models for neurodegenerative diseases are mainly transgenic mice based on genetic mutations, representing only 5% to 10% of these diseases, and have pathophysiological differences from humans, organoids can be an excellent facilitator for drug discovery [18,19]. Thus, we suggest that organoid-based precision medicine approaches can overcome the limits of animal models as a bridge between preclinical and clinical phases, translating applications from bench to bedside.
Combinatory systematic approaches with organoid-based omics analyses and in silico perturbation analyses
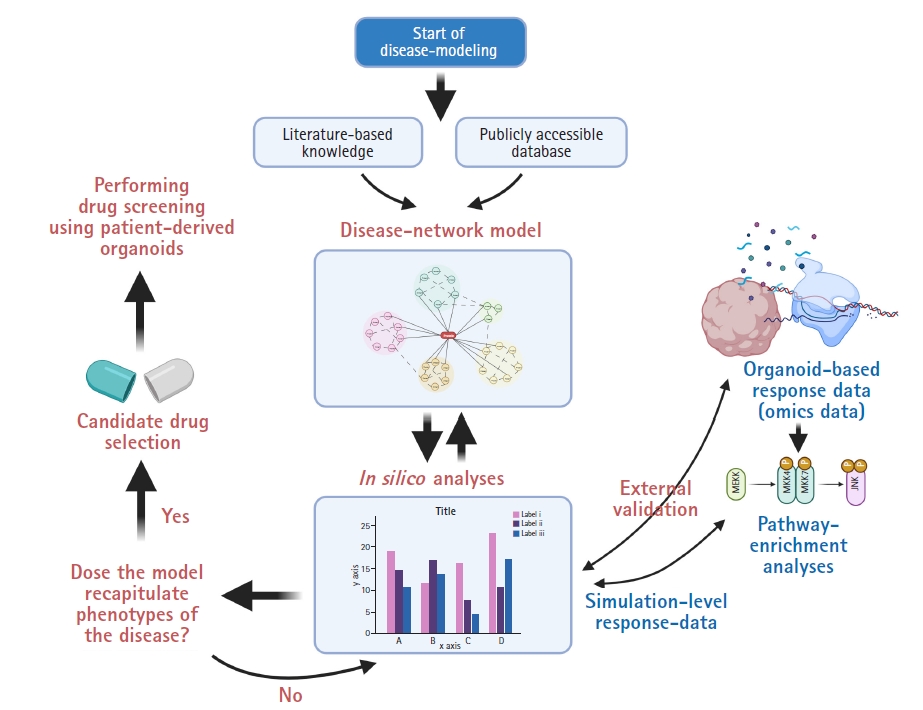
In recent years, there has been an enormous quantity of publicly available databases and knowledge. This has allowed researchers to recapitulate biological processes by systematically integrating polymorphic databases [20]. Although systems biology aims to understand the operation of complex biological processes and has enormous datasets to be explored, network models generated from these approaches require further in-depth validation using humanized samples, such as iPSC-derived organoids. In other words, the practicability of modulating molecular pathways inside a network model via medication needs individual validation because systems biology concentrates on complexity and emergent properties, not on external validation using individual biological samples [20–22]. For instance, large-scale data on the genomic, proteomic, or transcriptomic level from human organoids with information on the expression of genes or proteins can be compared with the results from in silico perturbation analyses of systems-biology-based disease network models [22]. In the field of neurological research, Park et al. [6] reported a mathematical network model for Alzheimer disease (AD) with the validation of in silico analysis using transcriptomic data from patient-derived brain organoids and developed a drug screening platform for precision medicine, Berger et al. [23] showed a computational simulation model of oxygen consumption using midbrain-specific organoids, and McMurtrey [24] investigated an equation-based in silico model to predict the development patterns of cerebral organoids. Fig. 2 shows strategies to develop a feedback loop integrating the construction of a network model and organoid-based omics datasets to validate the in silico analysis of candidate drugs.
Genetic modulation of patient-derived brain organoids
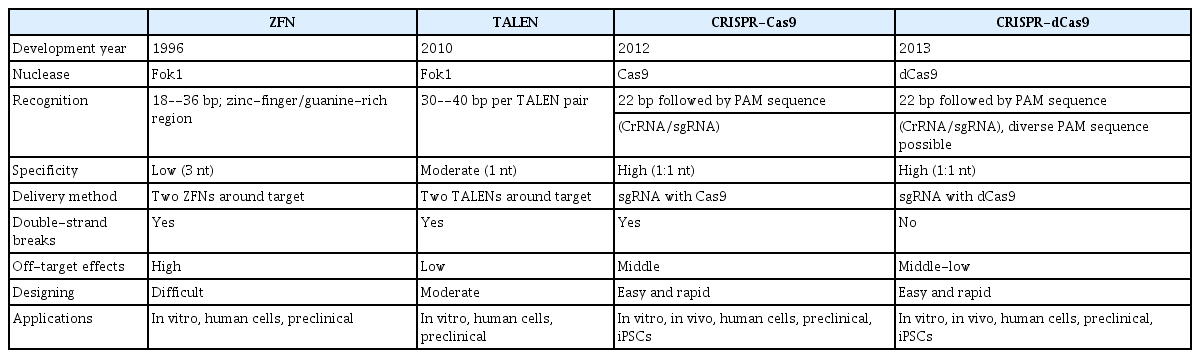
For polygenic neurodegenerative diseases such as AD, Parkinson’s disease, and amyotrophic lateral sclerosis, or other diseases with rare variants, the genetic engineering of organoids via zinc finger nucleases (ZFNs), transcription activator-like effector nucleases (TALENs), or CRISPR-Cas9 genome editing may be needed [25]. ZFN, an engineered DNA-binding protein, consists of two functional domains (the DNA-binding zinc finger domain and the DNA cleavage [Fok 1] domain) and facilitates genomic editing steps with double-strand breaks in DNA at specific locations [26,27]. For example, transfection of plasmid DNA including ZFNs can be used in human iPSCs to test the resistance of a specific gene to target drugs [28,29], and mutagenesis of the receptor for HIV by ZFN genome editing provided a functional cure for HIV/AIDS [30]. The TALEN system was developed in 2010–2011 and also has a DNA-binding domain and Fok I catalytic domain [31]. The monomeric components of the DNA-binding domain bind to each nucleotide of the target sequences and are responsible for the recognition of specific nucleotides. Although the TALEN system needs thymine before the end of the target sequence, current new techniques overcame this limitation through variants of the TALEN N-terminal domain [31,32]. The appearance of the CRISPR-Cas9 system was sensational due to its higher flexibility to target nucleotides and low levels of off-target effects [33,34]. The binding of the Cas9 nuclease to the protospacer-adjacent motif sequence helps in the direct induction of DNA double-strand breaks by the Cas9 protein [34]. More recently developed methods using catalytically dead Cas 9 (dCas9; a protein does not cleave DNA strands) and adenosine/cytidine deaminase induce the direct conversion of nucleotides without any double-strand DNA breaks [35,36]. These procedures must take precedence at the stage of embryonic stem cells or iPSCs in order to generate genetically engineered organoids, because the expansion of organoids is not possible in general. Assuming that there is a patient with a genetic mutation in a certain gene, current base-editing techniques can be utilized to substitute the inappropriate base pair in their patient-derived iPSCs. The engineered iPSCs can be used for generating organoids and then as a drug screening platform. Moreover, although current research has been restricted to human organoids-to-animal xenotransplantation due to their instability and ethical issues, it will be possible to conjugate organoids for organ transplants as an alternative route in the near future. Table 1 shows a comparison of the ZFN, TALEN, and CRISPR-Cas9 platforms, including current CRISPR techniques for base editing without double-strand DNA breaks [25,26,31,37–39].
Drug repositioning to accelerate clinical approval
Given the slow pace and high costs of new drug development, reconsidering previously approved drugs is increasingly becoming a fascinating approach to accelerate clinical approval [9,40]. Drug repositioning (also known as drug repurposing) is a novel experimental approach for investigating new possible uses for existing drugs beyond the scope of their original purposes [9]. Historically, most reported cases of drug repositioning, such as sildenafil or thalidomide, occurred accidentally instead of involving systematic approaches or omics-based research [40]. However, since many novel sources of data for drug repositioning have recently emerged and current techniques can simulate the effects of candidate drugs via in silico perturbation, drug repositioning “with a purpose” is becoming a promising way to explore alternative drugs [10]. Xu et al. [41] showed in silico drug repositioning using drug-virus, virus-virus, drug-drug similarity networks for coronavirus disease 2019. Park et al. [6] reported an in silico perturbation analysis using a mathematical network model for AD using FDA-approved candidate drugs, Cai et al. [9] introduced drug repositioning strategies based on a heterogeneous information fusion graph convolutional network, and Tran et al. [42] developed a prediction framework for drug-target interactions for drug repositioning with graph neural networks, called deep neural computation. Furthermore, recently developed advanced platforms provide numerous databases to guide scientists to valuable sources, such as Network-based Drug Repurposing and Exploration (NeDRex), ksRepo, and DrugNet [43–45]. Thus, assuming that there is a sufficient safety basis from phase I studies, the latest strategies for drug repositioning will enable us to enter phase II clinical assessments more efficiently and rapidly. Moreover, regarding organoid-based precision medicine, the clinical assessments can be much more successful because all samples are derived from human patients and treated with the selected candidate drugs via drug repositioning strategies.
Imaging and quantification techniques for drug screening using brain organoids
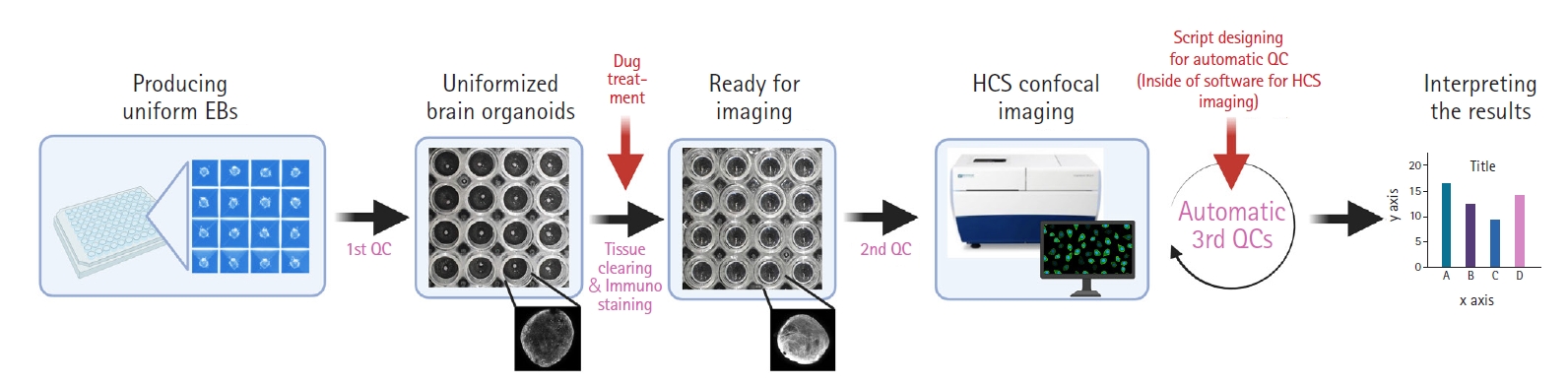
For the practical use of brain organoids as samples for drug screening, there are still many obstacles to outcomes. First, human brain organoids have a longer diameter (>1 mm) than 3D neuro-spheres (<300 μm) derived from neural stem cells and are entangled with a large number of cortical neurons. These traits of brain organoids make it difficult for antibodies to fully penetrate into their center. Fortunately, many scientists have developed cutting-edge protocols to tackle this issue. Woo et al. [46] have summarized current tissue-clearing methods well. There are three types of tissue clearing methods, as follows: (1) hydrogel-based tissue transformation, (2) clearing methods with hyperhydrating and high-refractive index aqueous solutions, and (3) clearing methods using organic solvents. Na et al. [47] also developed the sodium cholate-based assisted removal of lipid for fluorescent imaging of deep biological tissues method as a superior alternative to the sodium dodecyl sulfate-based methods. Second, although these methods enable us to visualize deep inside of brain organoids, simultaneous screening systems are necessary for effective drug assessments. Current advances in microscopic techniques have provided scientists with new perspectives on screening a large number of organoids, such as ImageXpress Micro Confocal (Molecular Devices, San Jose, CA, USA), CellInsight CX7 LZR (ThermoFisher, Waltham, MA, USA), and CellVoyager CV8000 HCS (Yokogawa, Tokyo, Japan) [48]. In particular, HCS systems with spinning disk confocal technology, such as the ImageXpress system, have made it possible to perform high content analysis with high-resolution images [6]. Third, automatic quality-control methods for organoids are urgently needed. Although recent culture devices for homogenous embryonic bodies, such as Aggrewell 800 (Stemcell Technologies, Vancouver, Canada) are well established, the remaining subtle morphological or size differences (physically unmanageable) can lead to poor results in drug evaluations. In the step of acquiring images, it would be important to designate commands or scripts to automatically detect size variations or morphological distortions. Fig. 3 shows a full set of guidelines for automatic drug screening platforms using the brain organoids.
Challenges, future directions, and conclusions
Although current brain organoids have been used as miniaturized human brains, the microglial population does not emerge during the developmental processes of brain organoids, due to their different origins in the mesodermal yolk-sac [49]. Since microglial cells are major immune cells in the brain, further studies using mixed cultures of iPSC-derived organoids and microglia are highly needed. Park et al. [13] demonstrated that a mixed-culture system is possible now and suggested the necessity of their advanced brain organoids with microglial cells, called “brain assembloids”. Next, the development of organoids with full vasculatures is also a great need for mimicking real human brains more accurately, although several researchers have developed human brain organoids with a functional vascular-like system (vhCOs) [50–53]. Cakir et al. [50] provided engineering methods for vhCOs, but they have some limits in terms of the fact that the vasculature of vhCOs is induced by a lentiviral infection containing FUW-tetO-ETV2, not spontaneously. Ham et al. [53] tried to generate endothelial cell (EC)-based vasculature under spontaneous conditions using vascular endothelial growth factor and confirmed EC markers such as CD31 and claudin-5; however, they did not show results of functional analyses or interactions between neurons and ECs. Moreover, no previous studies developing vasculature via iPSCs have considered the role of microglia. Taken together, future studies are needed in the context of the new era of organoid systems, including both microglia and vasculature, that can functionally interconnect each of the cell types. Third, organ-on-a-chip techniques incarnating the whole human body also can be a perspective for organoid-based research. For instance, recent studies have described the importance of the gut-brain axis, which consists of bidirectional interactions between the central nervous system and peripheral enteric nervous system [54,55]. In this respect, modeling the interaction of human brain organoids with intestine organoids will provide potential prospects for new in vitro biomimetic systems. Finally, the current formulaic approval process for clinical trials does not elaborate on the involvement of stem cell-derived organoid studies. For more advanced research and the materialization of organoid-based precision medicine, newly revised approval processes for stem cell-based organoid research will be demanded, with flexible but strict regulations for the effectiveness and safety of candidate drugs.
In this review, we discussed current methods for organoid-based precision medicine, limitations, and future perspectives. Fig. 4 shows a prospective drug screening system with possible applications of multi-organs-on-chip, HCS system, and systems-biological analyses. We herein suggest that organoid-based precision medicine approaches provide unique opportunities for neurodegenerative diseases, enable us to reduce the necessity of animal experiments, and maximize the efficacy of drugs as a bridge between preclinical and clinical phases, translating applications from the bench to bedside, and accelerate clinical approval steps in the future.
Notes
Conflict of interest
No potential conflict of interest relevant to this article was reported.
Funding
This work was supported by a National Research Foundation of Korea (NRF) grant funded by the Korean government (Ministry of Science and ICT, MSIT) (No. NRF-2022R1C1C2012736).
Author contributions
Graphical figures were created with BioRender.com by Jong-Chan Park, PhD.
Data availability
Please contact the corresponding author for data availability.