A brain metastasis model for breast cancer using human embryonic stem cell-derived cerebral organoids
Article information
Abstract
Background
Breast cancer is a common cause of brain metastasis. Although breast cancer has relatively high survival rates, its survival rate after metastasis to the brain is lower. Conventional two-dimensional cell culture models and animal models are widely used in metastatic cancer research, and these models have tremendously contributed to the understanding of this disease. However, these models have some limitations, such as different physiological features and genetic backgrounds.
Methods
We established a simple metastatic breast cancer model using human pluripotent stem cell-derived cerebral organoids (COs)—in this case, breast cancer cerebral organoids (BC-COs).
Results
Using the BC-CO model, we induced the metastasis of MDA-MB-231 cells into COs by co-culture of cells with COs and compared the differences between adapted cancer cells in BC-COs and non-adapted cells. Our results showed that the proliferative capacity increased in adapted cells. Additionally, the expression levels of endothelial-mesenchymal transition markers and cancer stem cells were significantly higher in adapted cancer cells.
Conclusion
We conclude that metastasis promotes the metastatic capacity of breast cancer cells. Our results also showed that the BC-CO model could be a novel tool for research on brain metastasis in breast cancer.
Introduction
Metastasis is a major feature of various cancers. Cancer cells move away from the primary site through the circulation and colonize secondary sites through complicated processes, such as adhesion, invasion, migration, and growth [1,2]. Breast cancer metastasis is the second most frequent cancer of the central nervous system [3]. Previously, it was reported that 30% to 50% of patients with metastatic breast cancer will develop brain metastasis in the course of their disease [4]. Triple-negative breast cancer (TNBC; negative for estrogen receptor, progesterone receptor, HER2/neu) has a poor prognosis compared to other breast cancer subtypes [5]. In particular, the rate of metastasis of TNBC within 5 years is 4 times higher than that of other types of breast cancer [6]. Despite aggressive treatments, including radiotherapy, chemotherapy, and surgery, the survival rate is low [7].
In vitro conventional cell culture models or orthotopic cancer xenograft mouse models are widely used as tools to study brain metastasis of breast cancer [8,9]. However, 2-dimensional (2D) cell culture models have obvious limitations, such as the poor replication of complex tissue structures and essential physiology shown in vivo, and the interspecies differences between humans and animals in physiological, genetic, and histological characteristics are also problems that need to be overcome [10–12]. In addition, animal experiments have the potential to cause several other problems, including ethical issues, inconvenient management, and high costs. Organoid technologies have been highlighted as alternative solutions. Organoids are 3-dimensional (3D) in vitro experimental models that accurately reflect the histological and physiological features of human organs [12–14]. The human brain is composed of various types of cells, including neurons, neuronal progenitors, astrocytes, oligodendrocytes, and microglia; this cellular diversity and these histological structures are well replicated in human pluripotent stem cell-derived cerebral organoids (COs) [15,16]. In this study, we established a simple model for brain metastasis of breast cancer using 70-day-old COs and MDA-MB-231 cells, a representative TNBC cell line. We investigated the changes that occur after metastasis of breast cancer cells using this model.
Materials and Methods
Ethics statement: This study was approved by the Public Institutional Review Board (No. P01-201910-41-001).
1. Human embryonic stem cell culture
All experiments involving human embryonic stem cells (hESCs) were approved by the Public Institutional Review Board (approval number: P01-201910-41-001) designated by the Ministry of Health and Welfare. The hESC line (SNUhES31) was obtained from the Institute of Reproductive Medicine and Population (Medical Research Center, Seoul National University Hospital, South Korea). hESCs were cultured on 10 μg/mL mitomycin-C (Roche, Mannheim, Germany)-treated mouse embryonic fibroblasts and maintained in an hESC medium comprising 20% knockout serum replacement (Life Technologies, Carlsbad, CA, USA), 1% minimum essential medium-non-essential amino acids (MEM-NEAA) (Life Technologies), 1% GlutaMAX (Life Technologies), and 7 μL/L β-mercaptoethanol (Sigma-Aldrich, St. Louis, MO, USA) in Dulbecco’s modified Eagle medium/nutrient mixture F-12 (DMEM/F-12; Thermo Fisher Scientific, Waltham, MA, USA) with 20 ng/mL basic fibroblast growth factor (bFGF; R&D Systems, Minneapolis, MN, USA). For feeder-free hESC culture, hESCs were detached from feeder cells using 1 mg/mL dispase (Life Technologies) and cultured in Essential-8 medium (Life Technologies) on Geltrex (Life Technologies)-coated culture plates. We subcultured the hESCs as small clusters every 4 days using a 0.5 mM ethylenediaminetetraacetic acid solution.
2. Cerebral organoid culture conditions
We generated COs from hESCs using a modified protocol [13]. Briefly, we dissociated feeder-free-cultured hESCs into single cells by incubation with Accutase (Life Technologies) for 15 minutes. Next, we plated the hESCs in 96-well U-bottom ultra-low attachment (ULA) plates (Corning, New York, NY, USA) at a density of 9×103/150 μL in an hESC medium containing 5 ng/mL bFGF and 50 μM Y27632, a Rho-associated protein kinase inhibitor (Tocris, Bristol, UK). We labeled this day as day 0. On day 2, the medium was replaced with fresh hESC medium containing 5 ng/mL bFGF and 50 μM Y27632. On day 4, the medium was changed to hESC medium without bFGF or Y27632. On day 6, we transferred each embryoid body (EB) to one well of a 24-well ULA plate containing 500 μL of neural induction medium comprising 1% N2 supplement (Life Technologies), 1% GlutaMAX, 1% MEM-NEAA, and 1 μg/mL heparin (Sigma-Aldrich) in DMEM/F-12. On day 8, the EBs were fed 500 μL of fresh neural induction medium. On day 10, we embedded the EBs in 20 μL of Matrigel (BD Bioscience, Franklin Lakes, NJ, USA) droplets and moved the Matrigel droplets into a 60-mm tissue culture dish (Corning) containing 5 mL of CO medium comprising 0.5% N2 supplement, B27 supplement without vitamin A (Life Technologies), 2.5 μg/mL of human insulin (Roche), 1% GlutaMAX, 0.5% MEM-NEAA, and 3.5 μL of β-mercaptoethanol in a 1:1 mixture of DMEM/F-12 and neurobasal medium (Life Technologies). The medium was changed every alternate day. On day 14, we transferred the Matrigel droplets containing COs to a 125-mL spinner flask with a CO medium comprising 0.5% N2 supplement, B27 supplement, 2.5 μg/mL human insulin, 1% GlutaMAX, 0.5% MEM-NEAA, and 3.5 μL of β-mercaptoethanol in a 1:1 mixture of DMEM/F-12 and neurobasal medium. The CO medium was changed every 7 days until 70-day-old COs were generated.
3. Establishment of an eGFP-expressing stable cancer cell line
The MDA-MB-231 cancer cell line was obtained from the Korean Cell Line Bank (Seoul National University, South Korea). These TNBC cell lines primarily arise from breast cancer. The cells were maintained in high-glucose DMEM supplemented with 10% fetal bovine serum (Gibco, Carlsbad, CA, USA) in a 5% CO2 humidified atmosphere at 37°C. For this study, we generated lentiviral constructs containing complementary DNA (cDNA) encoding enhanced green fluorescent protein (eGFP) and puromycin resistance genes. These transgene cassettes employed a cytomegalovirus (CMV) immediate-early promoter. The lentiviral and packaging vectors were kindly provided by Dr. Yibing Qyang (Yale Cardiovascular Research Center, Yale School of Medicine, USA). To construct CMV-eGFP-T2A-Puro, the relevant gene sequences were subcloned into a lentiviral vector. To generate CMV-eGFP-T2A-Puro lentiviral particles, we transfected the lentiviral and packaging vectors into HEK293T cells. The virus-containing medium was collected and concentrated by ultracentrifugation at 25,000 rpm for 2 hours (Hitachi Koki Co., Tokyo, Japan). Finally, we exposed MDA-MB-231 cells to a concentrated virus-containing medium for 24 hours, followed by culture for 2 days in a culture medium without the virus. To select virus-infected MDA-MB-231 cells, we treated cells with 10 μg/mL puromycin for 3 days.
4. Generation of breast cancer cerebral organoids and cancer cell adaptation
Breast cancer cerebral organoids (BC-COs) were generated by the implantation of breast cancer cells expressing eGFP on 70-day-old COs (Fig. 1). Briefly, we mixed each CO with 2×104 cancer cells in 200 μL of CO medium in a single well of a 96-well U-bottom ULA plate and then centrifuged them at 1,000 rpm for 3 minutes. The culture plates were incubated for 12 hours in a 5% CO2 humidified atmosphere. After the aggregation and adhesion of cancer cells to the CO surface (forming a BC-CO), we transferred each BC-CO into a single well of a 24-well flat-bottom ULA plate containing 500 μL of CO medium, followed by incubation for an additional 24 hours for stabilization. Finally, we moved the BC-COs into a 125 mL spinner flask containing CO medium. This day was set as BC-CO day 0, and culture continued until the analysis. Cancer cells on BC-COs at this time point were considered as first-adapted. To harvest the first-adapted cancer cells, the BC-COs were dissociated into single cells with Accumax (Stem Cell Technologies, Vancouver, BC, Canada), and eGFP-positive cells were sorted using BD FACS ARIA III (BD). To induce the second or third adaptation of cancer cells, we generated BC-COs by implanting first- or second-adapted cancer cells (2×104 cells) sorted by fluorescence-activated cell sorting (FACS) to new COs and cultured them for 7 days. The adapted cells were harvested using FACS.
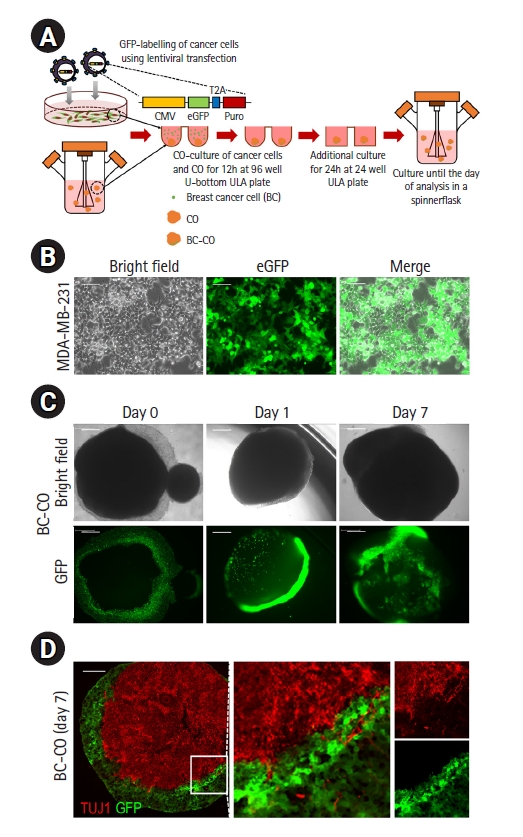
Construction of the BC-CO model. (A) Schematic of the protocol used to develop the BC-CO model. (B) Representative bright field and fluorescence images of eGFP expressing MDA-MB-231 cell lines infected with CMV-eGFP-T2A-Puro lentivirus. Scale bar=200 μm. (C) Representative bright field and fluorescence images of BC-COs at BC-CO days 0, 1, and 7. Scale bar=500 μm. (D) Representative images showing the coexistence of neurons in eGFP-positive cancer cell areas of BC-COs. Scale bar=200 μm. CO, cerebral organoid; BC-CO, breast cancer cerebral organoid; CMV, cytomegalovirus promoter; Puro, puromycin resistance gene; ULA, ultra-low attachment; eGFP, enhanced green fluorescent protein; TUJ1, neuron-specific class III-beta-tubulin 1.
5. Histology and immunofluorescence staining
We fixed COs or BC-COs in 4% paraformaldehyde (Junsei Chemical Co., Ltd., Tokyo, Japan) for 2 hours at 4°C and sequentially immersed the samples in 15% and 30% sucrose solutions until they sank. Next, the samples were embedded in an optimal cutting temperature compound (Leica, Wetzlar, Germany) and cryo-sectioned into 20-μm sections using a Leica CM1850 cryostat (Leica). For immunofluorescence staining, sections were permeabilized and blocked with 0.1% (v/v) Triton X-100 (Sigma-Aldrich) in phosphate-buffered saline (PBST) containing 10% normal goat serum (Vector Laboratories, Burlingame, CA, USA) for 1 hour at room temperature. The sections were then incubated with the following primary antibodies in PBST containing 2% normal goat serum at 4°C overnight: anti-neuron-specific class III beta-tubulin 1 (anti-TUJ1; rabbit, 1:100, Cell Signaling 5666). The sections were washed 3 times with PBST and then incubated with donkey Alexa Fluor 568 conjugate (1:500, Life Technologies) as a secondary antibody for 3 hours at room temperature:. Finally, the sections were again washed 3 times with PBST, mounted under coverslips on slides using Vectashield mounting medium (Vector Laboratories), and imaged using a TCS SP5 II confocal microscope (Leica).
6. Cell Counting Kit-8 assay
The cells were plated at a density of 1×104 cells/well in 96-well plates (BD Biosciences) and cultured. The Cell Counting Kit-8 (CCK-8) solution was added to each well at a 1:10 dilution, followed by further incubation at 37°C for 3 hours. Absorbance was measured at 450 nm using a microplate reader (BioTek Instruments Inc., Winooski, VT, USA).
7. Real‐time quantitative polymerase chain reaction
Total RNA was extracted using the RNeasy Plus RNA Extraction Kit (Qiagen, Hilden, Germany). Reverse transcription was performed using an iScript cDNA Synthesis Kit (Bio-Rad, Hercules, CA, USA). Reverse transcription products (2.5 ng cDNA) were amplified using a FastStart Essential DNA Green Master PCR Kit (Roche) and primers. The primers used here were as follows: 5′-GTTGGAGAAGGTGGAACCAA-3′ (forward) and 5′-CTCCTTCTGCAGGGCTTTC-3′ (reverse) for OCT4; 5′-TGG CGA ACC ATC TCT GTG GT-3′ (forward) and 5′-GGA AAG TTG GGA TCG AAC AAA AGC-3′ (reverse) for SOX2; 5′-CAGAAGGCCTCAGCACCTAC-3′ (forward) and 5′-ATTGTTCCAGGTCTGGTTGC-3′ (reverse) for NANOG; 5′- GGC ATA CAC CTA CTC AAC TAC GG-3′ (forward) and 5′- TGG GCG GTG TAG AAT CAG AGT C-3′ (reverse) for ZEB1; 5′-TTC CCA AAA AGA GGC TGA GA-3′ (forward) and 5′-CAA TGT TGC AAG GGT TTG TG-3′ (reverse) for CD44; 5′-AGG CAA AGC AGG AGT CCA CTG A-3′ (forward) and 5′-ATC TGG CGT TCC AGG GAC TCA T-3′ (reverse) for VIM. The samples were cycled 45 times using a LightCycler 96 Real‐Time System (Roche). Real‐time quantitative polymerase chain reaction (RT‐qPCR) was performed using the following conditions: 5 minutes at 95°C, 30 seconds at 95°C, 30 seconds at 58°C, and 30 seconds at 72°C. All RT‐qPCR experiments were performed in triplicate. The cycle threshold was calculated using default settings with real-time sequence-detection software (Roche).
8. Statistical analysis
Data are presented as mean ± standard error of the mean. The differences between mean values were analyzed using the Student t-test. A p-value <0.05 was considered statistically significant.
Results
1. Generation of the metastatic BC-CO model
We established the conditions for the generation of the BC-CO model (Fig. 1A). To distinguish breast cancer cells from BC-COs, we first generated an eGFP-labeled MDA-MB-231 cell line by CMV-eGFP-T2A-Puro lentiviral transfection, followed by selection with puromycin treatment. As shown in Fig. 1B, the selected cells strongly expressed GFP under 2D culture conditions (Fig. 1B). Next, we generated BC-COs by implanting eGFP-labeled cancer cells onto 70-day-old COs, and observed that cancer cells were attached to the surface of COs after implantation (day 0). After 7 days of culture in a spinner flask, we observed that the eGFP-labeled cancer cells proliferated significantly (day 7). Additionally, the cells appeared to move inside the CO (Fig. 1C). In BC-COs on day 7, immunohistochemical results showed the coexistence of TUJ1-positive cells and eGFP-positive cells at the interface of each cell population (Fig. 1D). These results indicate that cancer cells or neural cells migrated into each cell area in BC-COs, and there was a possibility of interaction between cancer cells and neural cells. Reciprocal interactions between cancer cells and neurons are a strategy for cancer cell survival, and cancer cells exploit neuron-derived factors to generate a positive microenvironment for survival and proliferation [17].
2. Effect of adaptation of breast cancer cells in BC-CO
In this study, we investigated the effects of adaptation of breast cancer cells in BC-COs. As shown in Fig. 2A, we observed an increase in the proliferative capacity of cancer cells as adaptation was repeated under a microscope (Fig. 2A). After the third adaptation, we harvested eGFP-positive cells by FACS sorting and cultured them in 2D culture conditions (Fig. 2B). Next, we compared the proliferative capacity of the third-adapted and non-adapted cells (Fig. 1B) using the CCK-8 assay. As shown in Fig. 2C, the proliferative capacity of third-adapted cancer cells was significantly higher than that of the non-adapted cancer cells. These results suggest that the characteristics of metastatic cancer cells may change dynamically.
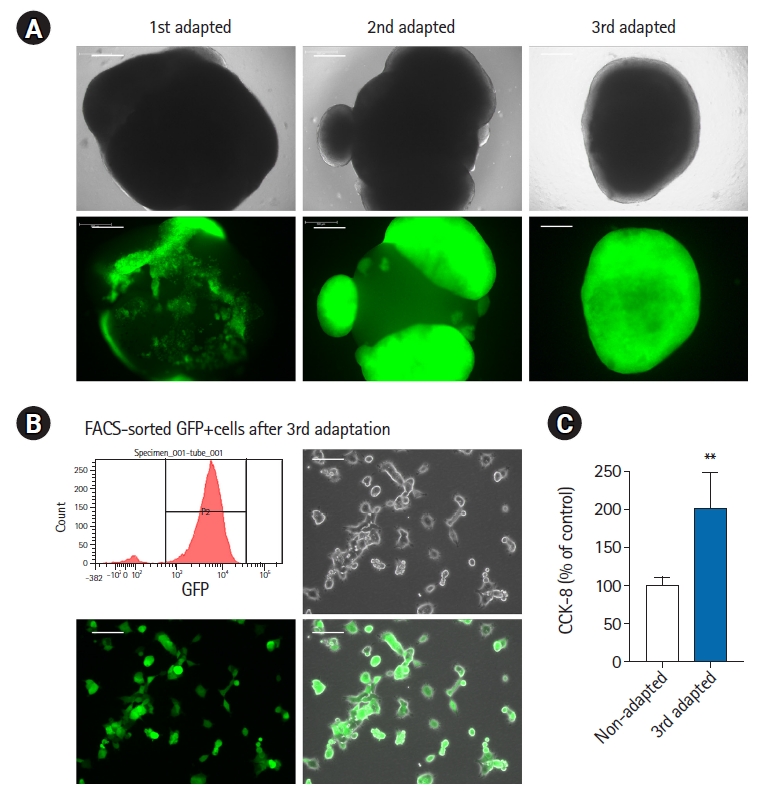
Adaptation of breast cancer cells in BC-COs. (A) Representative bright field and fluorescent images of BC-COs with first, second, and third-adapted MDA-MB-231 cells. Scale bar=500 μm. (B) Representative images of third-adapted eGFP-positive breast cancer cells sorted by FACS. Scale bar=200 μm. (C) Cell proliferation in non-adapted and third-adapted MDA-MB-231 cells was measured using a Cell Counting Kit-8 assay. **p<0.005. vs. non-adapted. BC-CO, breast cancer cerebral organoid; FACS, fluorescence-activated cell sorting; eGFP, enhanced green fluorescent protein.
3. Metastasis-related factors were increased after breast cancer adaptation
To observe the changes in metastasis-related markers in adapted cancer cells, we compared the expression of epithelial to mesenchymal transition (EMT) markers and cancer stem cell (CSC) markers between non-adapted and third-adapted cancer cells using RT-qPCR. As shown in Fig. 3A, the expression levels of EMT markers ZEB1 (zinc finger E-box binding homeobox 1), CD44 (HCAM, homing cell adhesion molecule), and VIM (vimentin) were significantly higher in third-adapted cancer cells than in non-adapted cells. Additionally, the expression levels of the CSC markers OCT4, NANOG, and SOX2 were also significantly higher in the third-adapted cells. These results indicate that the metastatic capacity of cancer cells increased after adaptation. Based on these results, we confirmed the possibility of using BC-COs as a useful tool to study brain metastasis in breast cancer.
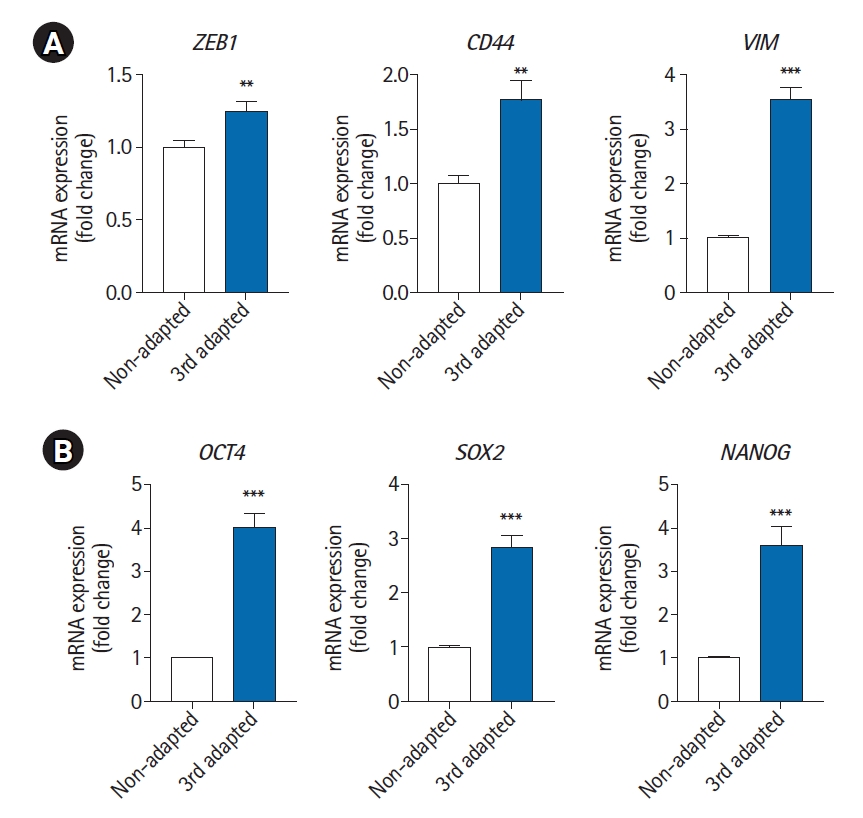
Expression levels of EMT and CSC markers in non-adapted and third-adapted MDA-MB-231 cells. The mRNA expression levels of EMT markers (ZEB1, CD44, and VIM) (A) and CSC markers (OCT4, SOX2, and NANOG) (B) were measured using real‐time quantitative polymerase chain reaction in non-adapted and third-adapted MDA-MB-231 cells. **p<0.005, ***p<0.001. vs. non-adapted. EMT, epithelial to mesenchymal transition; CSC, cancer stem cell.
Discussion
In this study, we established a new model for studying cancer metastasis using COs and BC-COs. Most studies on cancer biology are based on experiments using in vitro 2D cell cultures or animal models. However, 2D cell cultures have many limitations, such as the absence of natural tissue or tumor structures responsible for proliferation, gene expression, cell-cell or cell-microenvironment interactions, responsiveness to stimuli, drug metabolism, and other cellular functions [18–20]. Although orthotopic animal models have been used as a valuable tool to address host-tumor cell interaction issues, especially in metastatic cancer research, they also have limitations, such as interspecies differences, high costs, labor-intensive animal care, and ethical issues, which limit their utility in mechanistic studies [21,22]. These limitations can be addressed using the BC-CO model, which has various advantages. This model can be produced by the simple manipulation of breast cancer cells and COs. Because the BC-CO model is an in vitro approach, it has the advantages of time-dependent and real-time observations, the convenience of experimental manipulation, and precise physiological control. In addition, a functionally infinite number of samples with human genetic backgrounds can be obtained without ethical problems. In this study, BC-COs microscopically showed aspects of proliferation, invasion, and interaction with host cancer cells. Additionally, there were several differences between the 2D cultured cancer cells and the cancer cells in BC-COs. We found that the proliferative capacity of cancer cells in BC-COs significantly increased. Indeed, cancer cell proliferation is enhanced in brain metastasis through metabolic changes in cancer cells within the tumor microenvironment [23]. These results suggest that the BC-CO model reflects the features of metastatic cancer in the brain.
Recent research has shown that the EMT and CSCs play important roles in cancer metastasis [24]. The EMT is a process by which epithelial cells acquire the phenotype of mesenchymal stem cells [25]. This transition is involved in many fundamental processes, including embryonic evolution, tissue formation, wound healing, and tissue fibrosis, and it can promote tumor cell growth, drug resistance, and proliferation of tumors [25]. Importantly, a tremendous amount of research has established that the EMT plays a role in the metastasis of tumor cells [26,27]. CSCs represent a fraction of undifferentiated cancer cells that exhibit stem cell-like features and might represent a cellular resource that causes metastasis [28]. Additionally, the EMT enables the generation of CSCs during metastatic processes, including metastatic colonization [29]. In our results, breast cancer cells adapted to BC-COs exhibited increases in ZEB1, CD44, and VIM expression, which are known as EMT markers, and OCT4, SOX2, and NANOG, which are known as CSC markers [25,30]. It was previously reported that transcription factors are involved in the regulation of EMT in CSCs. For example, Wang et al. [31] showed that OCT4 and NANOG positively regulate the EMT and metastasis in breast cancer patients. In addition, another research group demonstrated that NANOG, by binding to the promoters of genes involved in EMT, including ZEB1, induces cancer metastasis [32]. Gao et al. [33] reported that SOX2 induces EMT and metastasis. These reports suggest that there is a close relationship between EMT and CSCs in regulating each other and their role in cancer cell metastasis. Therefore, our results indicate that BC-COs reflect changes in cancer cells during metastasis.
Metastasis is a complex process in which cancer cells disseminate from the primary tumor site and adhere to, proliferate, migrate to, or invade secondary sites [2]. Although it is difficult to reproduce the total process of cancer metastasis, the BC-CO model is a promising tool that provides additional information in conjunction with 2D or animal models as an in vitro model. Despite the potential of BC-COs, this model has many limitations, including a lack of brain vascular components or immune cells and data processing methods, which are important when considering metastasis data. Because novel organoid technologies are also being developed through diverse approaches, including genetic manipulation and cell and tissue engineering, these limitations will gradually be resolved.
Notes
Conflict of interest
No potential conflict of interest relevant to this article was reported.
Funding
This work was supported by National Research Foundation of Korea (NRF) grants funded by the Ministry of Science and ICT (No. 2020R1A5A2017323; No. 2021R1F1A1047379) and the fourth BK21 project (Educational Research Group for Platform Development of Management of Emerging Infectious Disease) funded by the Ministry of Education (No. 5199990614732) of the Korean government.
Data availability
Please contact the corresponding author for data availability.
