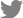Ethics statement: This study was approved by the Institutional Review Board of Asan Medical Center (No. #2018-0152).
1. Human specimens and organoid culture.
All samples used in this study were provided by the Asan Bio-Resource Center, Korea Biobank Network (2018-25(179)). The research protocol was approved by the Institutional Review Board of Asan Medical Center (#2018-0152; Seoul, Republic of Korea). The entire experimental protocol was conducted in compliance with the institutional guidelines.
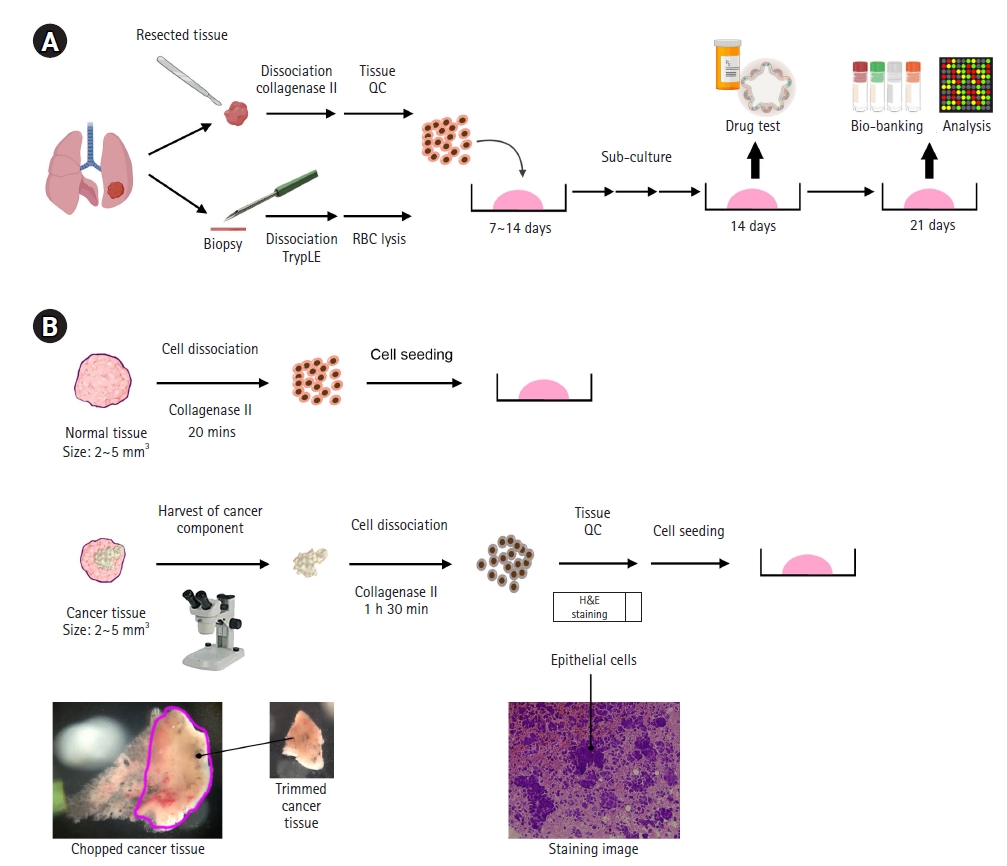
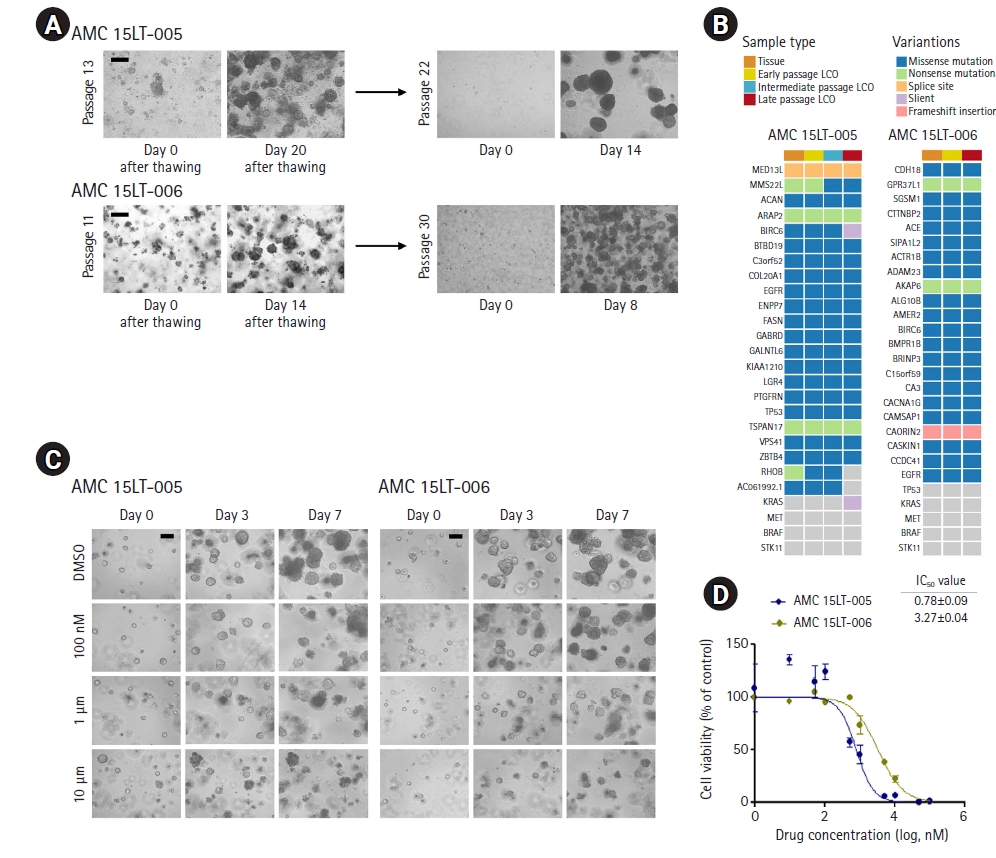
Surgically resected samples (approximately 1-4 cm3) and biopsy specimens (~1×30 mm) obtained from EBUS-TBNA were transported to the laboratory on ice within 1 hour of removal from patients in cold Hank’s balanced salt solution (HBSS) with antibiotics (Lonza, Basel, Switzerland). Samples were washed 3 times with cold HBSS that contained antibiotics. The surgically resected tissue was incubated with 0.001% DNase (Sigma-Aldrich, St. Louis, MO, USA), 1 mg/mL collagenase/dispase (Roche, Basel, Switzerland), 200 U/mL penicillin, 200 mg/mL streptomycin, and 0.5 mg/mL amphotericin B (2% antibiotics; Sigma-Aldrich) in DMEM/F12 medium (Lonza) at 37°C for 1 hour with intermittent agitation. The biopsy sample was incubated with TrypLE Express Enzyme (Gibco, Waltham, MA, USA) at 37°C for 5 minutes with intermittent agitation and treated with red blood cell (RBC) lysis buffer (Sigma-Aldrich) for 5 to 10 minutes at room temperature. After enzymatic digestion, the suspensions were repeatedly triturated by pipetting. The cells were centrifuged at 112×g for 3 minutes, and the pellet was resuspended in 50 μL of mininal basal medium (MBM) (serum-free medium (DMEM/F12; Lonza) supplemented with 20 ng/mL basic fibroblast growth factor (Invitrogen, Waltham, MA, USA), 50 ng/mL human epidermal growth factor (Invitrogen), N2 (Invitrogen), B27 (Invitrogen), 10 μM ROCK inhibitor (Enzo Life Sciences, New York, NY, USA), and 1% penicillin/streptomycin (Gibco) and 100 μL of Matrigel. The resulting cell suspension was allowed to solidify on pre-warmed 6-well culture plates (Corning, Corning, NY, USA) at 37°C for 10 minutes. After gelation, 3 mL of MBM was added to the well. The medium was changed every 4 days.
For passaging, a solidified Matrigel drop containing the organoids was harvested using cold Dulbecco’s phosphate-buffered saline (DPBS) and centrifuged at 1,000 rpm for 3 minutes at 4°C. The pellets were washed with cold DPBS and centrifuged at 1,500 rpm for 15 minutes at 4°C. The organoids were resuspended in 2 mL of TrypLE Express (Gibco) and incubated for 10 minutes at 37°C for dissociation. Later, 10 mL of DMEM/F12 containing 10% fetal bovine serum (FBS) was added, and the samples were centrifuged at 1,000 rpm for 3 minutes. The pellets were washed with DPBS and centrifuged at 1,000 rpm for 3 minutes. The pellets were resuspended in MBM+Matrigel (1:3) and reseeded at 1:3 to 1:4 ratios to allow the formation of new organoids.
To obtain stored LCOs, LCOs cultured in 3 to 4 wells of a 24-well plate were harvested using cold DPBS and then centrifuged at 1,000 rpm for 3 minutes at 4°C. The pellets were washed with cold DPBS and centrifuged at 1,500 rpm for 15 minutes at 4°C. The supernatant was removed and the LCO pellet was resuspended in freezing medium: Culture Media 7: ES grade FBS (Gibco) 2: DMSO (Sigma-Aldrich) 1. The vials for storage were transferred to a deep freezer, followed by a gradual reduction in temperature to −80°C, and transferred to vapor-phase liquid nitrogen storage.
3. Genomic analysis
DNA was extracted from tumor tissues with matched normal tissues and early-, intermediate-, and late-passage LCOs from AMC 15LT-005 and 006 lines using the DNeasy Blood & Tissue Kit (Qiagen, Hilden, Germany) according to the manufacturer’s protocol. In order to evaluate the somatic mutations in the paired tumor tissues and early-, intermediate-, and late-passage LCOs, Whole-exome sequencing was performed using the Illumina Hiseq 2500 platform. A SureSelect Exome Enrichment kit version 6.0 (Agilent Technologies, Santa Clara, CA, USA) was used to capture all exome regions for whole-exome sequencing. The DNA libraries were sequenced for 150-bp paired-end sequencing. The Illumina sequencing data were processed using the Genome Analysis Toolkit (GATK) v1.6.5 [
6]. The sequenced reads were aligned to the human reference genome (NCBI build 37) with the Burrows-Wheeler Aligner (ver. 0.5.9) [
7] using the default options. De-multiplexing was performed using MarkDuplicates of the Picard package to remove polymerase chain reaction duplicates. De-duplicated reads were realigned at known indel positions with the GATK IndelRealigner, and the base quality was recalibrated using the GATK TableRecalibration [
6,
8]. Somatic variant calling for single-nucleotide variants and short indels was performed with matched normal tissues using MuTect (v1.1.7) [
9] and SomaticIndelocator in GATK, respectively. Germline variants from somatic variant candidates were filtered out with common dbsnp (build 141; found in ≥1% of samples), the Korean Reference Genome Database, and a panel of normal samples. The final somatic variants were annotated using the Variant Effect Predictor (v79) and then converted to a mutation annotation file format using vcf2maf. Quality checks for fastq files were performed using FastQC (
http://www.bioinformatics.babraham.ac.uk/projects/fastqc/). Quality checks for analysis-ready BAM files, including the total read number, percentage of pass-filter reads, percentage of selected bases, mean target depth, percentage of target not covered, percentage of target bases covered 30X, and the percentage duplication for cleaned BAM files was performed using the CalculateHSMetrics and MarkDuplicates modules of the Picard program. Integrative Genomics Viewer was used to view the BAM files [
10]. R software (v3.0) with the maftools [
11], ComplexHeatmap, and ggplot2 packages were used for data visualization. Fingerprinting analysis was performed using the panel detecting 24 SNP alleles on the Sequenom MassARRAY technology platform (Sequenom, CA, USA).
4. Drug screening
LCOs cultured in 6 to 12 wells in a 24-well plate over 2 weeks were harvested and dissociated using TrypLE Express (Gibco). The dissociated LCOs were counted and seeded to 10 μL of 2×103 cells per well on a 96-well white plate (Corning) after being mixed in MBM+Matrigel (1:3 ratio). After gelation, 100 μL of MBM was added to each well. The LCOs were allowed to grow to a diameter of 60 to 70 μm. Next, gefitinib (Selleck Chemicals, Houston, TX, USA) was administered in 10 concentration ranges every 3 days in triplicate. DMSO treatment was applied as a control. After 6 days, the medium was changed to 100 μL of MBM per well to measure cell viability, and 100 μL of CellTiter-Glo (Promega, Madison, WI, USA) was added to each well. The plates were agitated for 30 minutes at room temperature prior to luminescence reading. IC50 values were determined using GraphPad Prism (v5.01; GraphPad LLC, San Diego, CA).








 PDF Links
PDF Links PubReader
PubReader ePub Link
ePub Link Full text via DOI
Full text via DOI Download Citation
Download Citation Print
Print